Translate this page into:
Identification of novel bacteriocin against Staphylococcus and Bacillus species
Address for correspondence: Abdu Aldarhami, Department of Medical Microbiology, Qunfudah Faculty of Medicine, Umm Al-Qura University, Al-Qunfudah, Saudi Arabia. Phone: +00966550035527. E-mail: ahdarhami@uqu.edu.sa
This is an open-access article distributed under the terms of the Creative Commons Attribution-Noncommercial-Share Alike 3.0 Unported, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
This article was originally published by Qassim Uninversity and was migrated to Scientific Scholar after the change of Publisher.
Abstract
Objectives:
Due to the continues emergence of antimicrobial resistance, discovery of novel compounds are urgently required. Thus, this study is focused to identify a novel antimicrobial peptide (bacteriocin) targeting multidrug-resistant pathogenic bacteria.
Methods:
About 80 environmental isolates were recovered and screened for anti-bacterial activity using simultaneous antagonism assays. Produced peptide (AB3) was purified using Strata-XL-C and Sep-Pack columns. Matrix-assisted laser desorption/ionization-time of flight (MALDI-TOF) analysis and MIC were conducted on the AB3 peptide to determine its molecular weight and spectrum of activity. Extraction and amplification for the 16S rRNA gene of the producing strain was accomplished using QIAamp DNA Mini Kit and GeneAmp PCR system thermocycler, respectively. Novelty of the compound was assessed based on all obtained genomic and proteomic data using Basic Local Alignment Search Tool search and Unni-Prot and Bactibase, respectively.
Results:
About 5% of screened isolates showed antagonistic activity toward tested indicators. Obtained compound showed narrow spectrum of activity toward certain Gram-positive species including, methicillin-sensitive Staphylococcus aureus (MSSA), methicillin-resistant Staphylococcus aureus (MRSA) and Bacillus species. MALDI-TOF analysis revealed the flowing molecular masses: 1288.207 Da, 1304.536 Da, 1326.529 Da, 1403.591 Da, and 1472.792 Da. The extensive genomic and proteomic analysis have indicated the discovery of novel bacteriocin produced by Bacillus malacitensis.
Conclusion:
A novel bacteriocin (AB3) was identified from B. malacitensis, which has showed promising in vitro bactericidal activity toward MSSA, MRSA, and Bacillus subtilis. This compound holds great potential to replace or used in combination with currently used antibiotics to treat serious untreatable bacterial infections. However, further investigations to determine its suitability for therapeutic use in human health are needed.
Keywords
Antimicrobial discovery
antimicrobial peptides
bacteriocins
methicillin-resistant Staphylococcus aureus
Bacillus malacitensi
Introduction
It is well-known that antimicrobial resistance (AMR) is a global growing crisis carries the greatest threat to human health.[1] At present, antibiotic drugs are claimed as the most effective therapeutic choice for bacterial infections in human health.[1-4] In addition, almost all modern medicine procedures, including heart surgeries, hip-replacements and even caesarean operations, are heavily relent on the use of antibiotics.[4,5] Furthermore, infections caused by drug resistant bacteria can lead to serious health and economic burdens nationally and worldwide.[6] This is because the high possibility of long hospitalization, recurrent infections, and increased rate of morbidity and mortality.[7] For instance, infections caused by multidrug-resistant (MDR) organisms, such as strains of Acinetobacter baumannii, Klebsiella pneumoniae, and Pseudomonas aeruginosa, are extremely serious and life-threating condition due to the difficulty of treatment and high rate of fatality as a consequence. In 2016, it was estimated that the AMR would lead to 10 million death annually by 2050, unless multiple actions to tackle the AMR would be successfully implemented before this date.[8-10] Consequently, searching for novel antibiotic as well as antibiotics-alternative therapies, including vaccines, antibodies, and bacteriocins are highly and urgently required.[11-13]
Bacteriocins are a group of antimicrobial peptides (AMPs) that are ribosommaly synthesized (naturally) by many bacteria and certain Archea.[14-16] Bacteriocins are the most promising class of AMPs to replace antibiotics drugs or used in combination with currently used therapeutic drugs. This is because bacteriocins are able to exhibit broad-spectrum antibacterial activity, great stability to acids and high temperatures, reduced probability of resistance, simplicity of production as well as absent or limited toxicity to human cells.[13-18] Produced bacteriocin could be used by the producing strain to fight other microbial strains or species for the nutrition or/and limited space in natural ecological niches.[19,20] Activity of bacteriocin is mainly toward closely-related bacterial species of the producer strain, but infrequently act against unrelated species.[21] Producer strains protect themselves from the activity of the produced compound (bacteriocin) through an immune protein and/or a sensing protein which controls either the synthesis or transportation of the active toxin.[22,23] Bacteriocins are working by multiple mode actions including, pore formation into the cell membrane of sensitive microorganisms, inhibition for the formation of bacterial cell wall and inhibition for the activity of RNase or DNase enzymes.[19]
There are various types of bacteriocins depends on their size, mode of action, microbial target, and production mechanism.[13] However, generally, bacteriocins can be divided into two groups based on Gram-stain type of the producing bacterium as bacteriocins of Gram-positive bacteria and bacteriocins of Gram-negative bacteria. Nevertheless, bacteriocins of Gram-positive bacteria can be categorized into four separate classes depending on its molecular weights, chemical structures and stability to high temperature, acids, and enzymes. Class I, known as lantibiotics or lanthionine-containing bacteriocins, consists of very small peptides (<5 kDa) that are thermostable. The best-known example of this class is nisin, which is produced by certain strains of Lactococcus lactis sub-species and is the most globally-used bacteriocin as a preservative in food products. Nisin is able to inhibit the germination of clostridial spores (e.g., Clostridium botulinum in pasteurized processed cheese), which prevents the formation of its toxins. Class II comprises larger peptides (<10 kDa), which are thermostable and possess an amphiphilic helical structure that permits their accession to the cytoplasmic membrane of the target cells resulting in depolarization of the membrane and cell death. This class of bacteriocins can be further divided into three subclasses: subclass IIa, that includes bacteriocins with high specificity towards Listeria monocytogenes(e.g., pediocin PA-1); subclass IIb usually refers to bacteriocins that are made of two peptides acting in combination and showing synergistic effect and subclass IIc includes certain peptides, such as circularin A and reutericin6. Class III comprises large thermolabile peptides (>30 kDa), which are complex in terms of its activities and protein structures. Finally, bacteriocins of large molecular weight and containing either carbohydrate or lipids moieties are classified as class IV.[11,13,24,25]
Multiple extensive studies have yielded hundreds of novel bacteriocins that act against various microorganisms, including drug resistant organisms.[13] For instance, nisin P, which was isolated from Streptococcus gallolyticus strain AB 39, acts against both methicillin-resistant Staphylococcus aureus (MRSA) and vancomycin-resistant Enterococci.[26] This study aims to identify a novel AMP active against clinically relevant bacteria. Purification and further investigations of the identified peptide, such as matrix-assisted laser desorption/ionization-time of flight (MALDI-TOF) analysis, minimum inhibitory concentration (MIC) and minimum bactericidal concentration (MBC) were performed to determine the novelty of the peptide and to evaluate its suitability for potential therapeutic applications, including potential use as antibacterial drug treating bacterial infections caused by MDR bacteria in human medicine.
Materials and Methods
Samples collection and bacterial isolation
About ten environmental samples (water, soil, and sewage) were collected using Falcon tubes (Corning incorporated, Corning, NY, USA) from different locations to ensure the diversity of potential bacteria. All solid samples, such as soil, were mixed thoroughly and were then diluted using distilled water, while aqueous samples, water, for example, were just mixed thoroughly. About four inoculated blood agar plates for each tested sample were incubated at different growth conditions in relation to temperature and oxygen availability to ensure the maximum rate of recovery. These conditions include aerobic and anaerobic incubation at 4°C, 16°C, laboratory temperature (LT), 30°C, 37°C, and 37°C in an aerobic atmosphere supplemented with 5% carbon dioxide (CO2).
Antagonistic testing
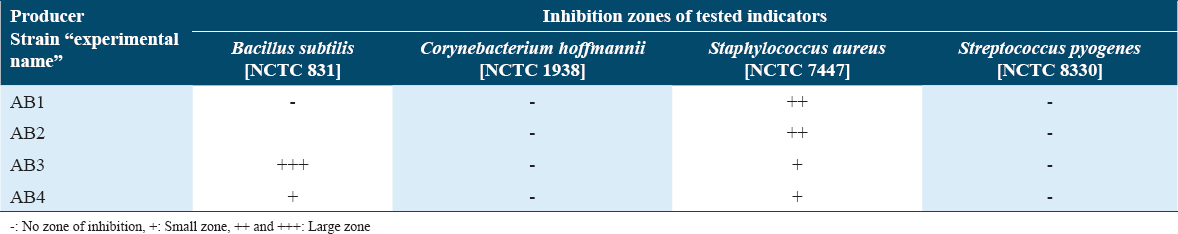
To detect potential antimicrobial producing strain/s simultaneous antagonism assays were performed.[27] Observed zones of inhibition on the lawn of the sensitive bacteria were measured using an arbitrary unit scale, in which the smallest zone scored “+” while “++++” was assigned for the largest zone of inhibition. Multiple reference and local indicator strains were used throughout this experiment for both Gram-positive and negative bacteria [Table 1]. Susceptibility profiles of local indictors were determined according to the British Society of Antimicrobial Chemotherapy (BSAC) standards.[28] First, 0.5 McFarland suspensions were prepared from obtained indicators and diluted to either 1:10 or 1:100 according to the BSAC standards.[28] Each Indicator was then lawn inoculated on a single nutrient agar medium (Merck, USA), followed by stab inoculating all recovered bacteria, which was identified based on their colonial morphology, using sterile wooden toothpicks. Plates inoculated with Gram-positive indicators were incubated for 18 h at 37°C plus 5% CO2, while those inoculated with Gram-negative indicators were placed in 37°C incubator for 18 h. Temperature of the incubation at this stage is highly dependent on the optimum temperature of both the recovered bacteria and indicator strains. Detected producer strains were evaluated by measuring zones of inhibition to choose the most appropriate producer for further investigation. A producer strain that acts against Bacillus subtilis (NCTC 831) and Staphylococcus aureus (NCTC 7447) was chosen for further investigation, and thus it was sub-cultured on a Columbia Blood Agar (CBA) after it was given an experimental name, which was AB3. In addition, B. subtilis was then selected as the indicator/sensitive strain throughout the investigation of the produced AMP from AB3 strain.

Broth optimization for AMP production
To enable high yield of the potential peptide from AB3 strain, this producing strain was inoculated on Tryptone Soya Broth with and without 5% of certain additives (glucose, lactose, and sucrose [Merck, USA]). Then, each prepared broth media (20 mL) was seeded with 5% (1 mL) from 0.5 McFarland suspension of the AB3 strain. Next, inoculated broth media were incubated at 37°C for 18–22 h using a shaker incubator (200 rpm) (New Brunswick Scientific, USA) to obtain oxygen very easily. Detection for the successful production of the potential compound and its overall yield was determined by performing multiple well diffusion assays against the sensitive strain (B. subtilis).
Well diffusion assay
Well diffusion assay was used to assess successful production and overall yield of the potential peptide in broth media as described by Tagg and McGiven in 1971.[29] About four wells were generated in each CBA using a sterile 4 mm cork-borer, and the bottoms of these wells were then sealed by adding few drops of a low melting temperature agarose gel (Merck, USA) to ensure the successful diffusion of the peptide into the blood agar. Supernatant from each inoculated broth media, which was obtained when about 1 mL of each positive broth media was centrifuged at 13,000 rpm for 5 min, was added its designated well. When the supernatant was completely diffused into the CBA, sterilization of the plates was performed by a direct exposure to chloroform vapor (Fisher Scientific, USA) for 10–15 min in a fume cabinet. This was performed to eliminate all viable bacteria including the producer strain and other contaminants to ensure that any observed inhibitory zone shall only reflect the activity of the peptide. Finally, the indicator strain, B. subtilis, was diluted and lawn inoculated on all CBA plates which were then incubated at 37°C plus 5% CO2 for 18 h. Zones of inhibition was monitored and measured to identify the best broth media, which made the highest yield of the potential peptide.
Spot inhibition assay
A spot inhibition assay is mainly performed to determine the activity of the AMP from supernatants or chromatography fractions, although it’s not commonly used for measuring zones of inhibition. A 1:10 dilution of B. subtilis was prepared and was then lawn inoculated onto CBA plates. Then, 10 μL of the sample (supernatants or chromatography fraction) was spotted on the CBAplate that was left to dry at LT. CBA Plate was ten sterilized using chloroform sterilization as mentioned above, followed by overnight incubation at 37°C plus 5% CO2.
Strata-XL-chromatography
To determine the molecular mass and any further investigation for the isolated AMP, the crude extract was initially purified by using Strata-XL-C (Phenomonex Ltd., USA), which was based on cation exchange. A volume of 10 mL (5 %) of 0.5 McFarland suspension of the AB3 producer strain was added into 200 mL of TSB (optimum media) in a conical flask. The flask was then placed into a shaker incubator at 37°C and 200 rpm for 18–22 h. Next, the broth was centrifuged at 13,000 rpm for 10–15 min and the supernatant was then collected and adjusted to pH 6.0. The Strata column was conditioned by adding 40 mL of acidified 90% methanol (pH 2) (Fisher Scientific Ltd., USA) followed by 80 mL of pure methanol to clean any previously bound peptide or impurities from the column. Before loading the supernatant, the column was washed by 40 mL of ultra-pure water to remove any remaining methanol, which might prevent the peptide from binding to the column. When the supernatant was loaded, 40 mL of 50% methanol was added (first elution), followed by a 60 mL of acidified 90% methanol (pH 2) (second elution). Active fraction was detected using the above-mentioned spot inhibition assay.
Sep-Pack C18 chromatography
The active fraction from Strata purification was further purified using a Sep-Pack column (Waters, USA). First, methanol was evaporated by placing the sample in a water bath at 75°C for 8 h. The sample was then centrifuged at 13,000 rpm for 5 min and 1 mL of the supernatant was diluted by adding 1 mL of ultra-pure water to enable easy flow of the sample through the column. The sample was then acidified to pH 2 by adding 0.1% of a trifluoroacetic acid (Merck, USA) to enhance the binding of the peptide to the column. The Sep-Pack column was conditioned by adding 2 mL of 100% acetonitrile plus 0.1% of a trifluoroacetic acid (buffer A) to remove any previously bound peptide or impurities from the column. Moreover, 2 mL of ultra-pure water acidified with 0.1% of the trifluoroacetic acid (buffer B) was added to wash out any residual acetonitrile, which might disturb the binding of the peptide. The sample was loaded into the column, followed by adding 2 mL of buffer B to remove unbound peptide. About 30 mL of air (empty syringe) was passed through the column to flush out any impurities from the column. Acetonitrile gradient from 0-100% was used to elute the peptide. Active fraction including the peptide was detected by performing spot inhibition assays for all fractions. All active fractions were combined and used as partially pure AMP preparation.
MALDI-TOF mass spectrometry analysis
MALDI-TOF analysis was performed on the active and purist fraction of the peptide to determine its molecular mass. Before conducting MALDI-TOF analysis, acetonitrile was removed from the active fraction using a rotary evaporator (Savant, USA), which was sent for MALDI analysis. The obtained molecular mass of the peptide was then searched against Bactibase database [http://bactibase.pfba-lab-tun.org/main.php] and protein UniProt [https://www.uniprot.org/uniprotkb?query=*] to determine its novelty.[30,31]
Since the active fraction from the Sep-Pack purification was further concentrated and purified, it was worth testing the activity of this fraction again to evaluate the spectrum of its activity. About 100 μL of the purified peptide was used to test its activity against all previously mentioned indicator strains [Table 1] as well as Bacillus cereus [NCTC 6349] and MRSA [University strain 393]. This was accomplished by carrying out well diffusion assays.
Determination of the MIC
The MIC for the AB3 peptide was carried out according to the protocol set out by Clinical and Laboratory Standards Institute (CLSI) (http://www.clsi.org). First, 0.5 McFarland suspensions of methicillin-sensitive Staphylococcus aureus (MSSA), MRSA, and B. Subtilis were prepared and then were diluted to 1:10. After a single row of 12 wells of a micro-titer plate was assigned to each indicator, 100 μL of Mueller Hinton broth (Merck, USA) was added to all 12 wells in each row. A 100 μL of the peptide solution was adjusted to pH 7 and was added to the first well of each row. Serial dilution (2-folds) was carried out using a multi-channel pipette on all wells apart from well 12, which was left as a growth control. Next, 5 μL of the prepared inoculum of each indicator was added to its designated wells. The micro-titer plate was then incubated overnight at 37°C.10 μL of the growth control of each indicator was diluted in 10 mL of saline and mixed thoroughly. Then, a 100 μL aliquot was spotted on Mueller-Hinton agar (MHA) and incubated overnight at 37°C. After incubation, colonies count was performed to calculate the original inoculum. In addition, 10 μL of the peptide solution was lawn inoculated on MHA to check its sterility. All plates were then incubated at 37°C for 18 h.
Determination of the MBC
To determine the MBC, clear wells (no visible bacterial growth) from the MIC experiment of each sensitive bacterium were serially diluted. 180 μL of ultra-pure water was added to 8 wells in each row of a new micro-titer plate. This was followed by adding 20 μL of each clear well from the MIC experiment to the first well of each row. Next, serial dilution was performed on all 8 wells of each row. Then, about 5 μL from each dilution belongs to each sensitive bacterium was spotted on MHA and incubated at 37°C for 18 h. The number of colonies of each dilution was recorded and compared with that of the original inoculum.
16SrRNA gene extraction and sequencing
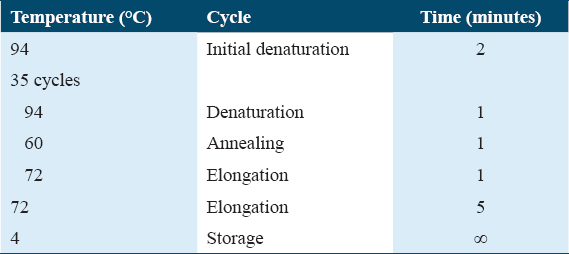
The antimicrobial producing strain (AB3) was identified via 16S rRNA gene sequencing. Gram-staining for the AB3 producer strain was performed to identify the most appropriate method for DNA extraction. About 5 McFarland suspension of the AB3 producer was prepared and the DNA was extracted using QIAamp DNA Mini Kit (Qiagen, UK) and lysozyme, lysostaphin, and mutanolysin enzymes (Merck, USA) on a heating block at 37°C for 30 min. Polymerase chain reaction (PCR) was carried out using GeneAmp PCR system 9700a thermocycler (Applied Biosystems, USA) and according to the parameters shown in Table 2. The PCR product was then run into an Invitrogen E-gel 2% agarose (Life Technologies, USA). Next, the PCR product was cleaned up using EXO SAP-IT-PCR Clean-up Kit (Affymetrix, USA) and was then sequenced using universal primers (Eurofins, Germany). The sequence of the forward primer was 63f (5’-CAG GCC TAA CAC ATG CAA GTC-3’), while 1387r (5’-GGG CGG WGT GTA CAA GGC-3’) belongs to the sequence of the reverse primer. The concentration of both primers was 10 pmol/μL. The sequencing was carried out using a 3730 48-capillary Genetic Analyzer (Applied Biosystems, USA). Obtained sequencing data were analyzed using BioEdit software and searched against the National Center for Biotechnology Information (NCBI) database using Basic Local Alignment Search Tool (BLAST) search to find out the identity of the AB3 producer strain.
Results
Recovered bacteria
Each collected environmental sample was inoculated into four growth conditions using CBA plates to increase the recovery rate of environmental bacteria. This isolation was resulted in numerous different bacteria depending on their colony morphology (size, color, hemolysis, edge, elevation, and surface) [Figure 1], and a total of eighty different strains were screened for antimicrobial activity.
- Examples demonstrating numerous bacteria recovered from environmental samples; (a) soil sample collected from underneath a rubbish bin (b) sample of sea water
Detected and selected antimicrobial producing strain
Antagonism test was performed by stab-inoculating all recovered bacteria on a CBA that was previously inoculated by a single indicator strain [Table 1]. About four antimicrobial producer strains, that mainly showed activity against Gram-positive indicators, were detected [Table 3]. Antagonistic activity toward both B. subtilis and S. aureus indictor strains was observed by AB3 and AB4, although bigger size of inhibitory zone was monitored for AB3 producing strain toward B. subtilis. Other two producing strains (AB1 and AB2) were only active against S. aureus indicator strain. There was no antimicrobial activity observed towards all tested Gram-negative indicators (data not shown). AB3 producer strain was selected for further study since it has exhibited a clear and large zone of inhibition against B. subtilis.
Broth optimization for the production of the peptide
Well diffusion assays were carried out on all used broth media for the growth of AB3 strain to determine the best medium, which has enabled the highest yield of the peptide. Size of inhibitory zones caused by the produced peptide after the growth of the AB3 producer into various broth media was measured, in which TSB without any additives [Figure 2] has showed the clearest and largest zone of inhibition (9 mm).
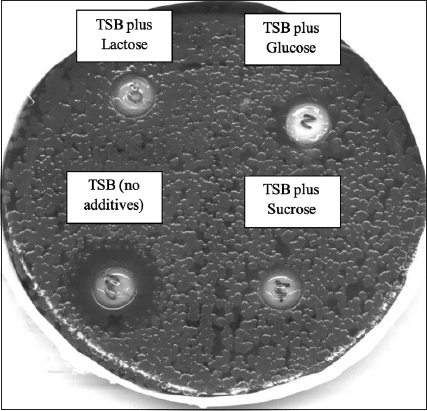
- Well diffusion assay showing the activity of the produced peptide against Bacillus subtilis after growing the producing strain (AB3) into a Tryptone Soya Broth with and without the addition of 5% of certain sugars (glucose, sucrose, and lactose)
Purification and MALDI-TOF analysis of AB3 peptide
During Strata purification, the active fraction was eluted by acidified 90% methanol (pH 2), whereas 90 and 100% acetonitrile was used to elute the peptide during the Sep-Pack chromatography. Size of inhibitory zone caused by the peptide against B. subtilis after Strata purification was 11 mm, whereas a 14 mm zone of inhibition was recorded for the peptide after the Sep-Pack chromatography. The result of MALDI-TOF analysis showed five peaks with different molecular masses at 1288.207 Da, 1304.536 Da, 1326.529 Da, 1403.591 Da, and 1472.792 Da.
Spectrum of activity
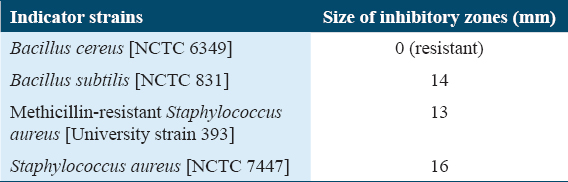
After all successful steps of purification for the AB3 peptide, its spectrum activity is mainly towards Gram-positive bacteria, although tested strain of B. cereus [NCTC 6349] was resistant [Table 4]. In addition, all tested Gram-negative indicators were fully resistant (data not shown).
MIC and MBC of AB3 peptide
As shown from Figure 3, the MIC for both B. subtilis and MRSA was observed at well 3 (1/8 of the undiluted peptide), while well 4 (1/16 of the undiluted peptide) reflected the MIC for MSSA. With the respect of the MBC, there was no bacterial growth (colonies) observed in all dilutions of B. subtilis, MRSA, and MSSA after 18 h of incubation.
![The minimum inhibitory concentration of the AB3 peptide against Bacillus subtilis [NCTC 831], methicillin-sensitive Staphylococcus aureus ([MSSA] NCTC 7447) and methicillin-resistant Staphylococcus aureus ([MRSA] University strain 393)](/content/195/2023/17/5/img/IJHS-17-15-g007.png)
- The minimum inhibitory concentration of the AB3 peptide against Bacillus subtilis [NCTC 831], methicillin-sensitive Staphylococcus aureus ([MSSA] NCTC 7447) and methicillin-resistant Staphylococcus aureus ([MRSA] University strain 393)
Identity of the AB3 producer strain
To identify the producer strain, the 16S rRNA gene of the producer was sequenced. When searching the obtained sequences of AB3 16S rRNA gene against the NCBI database using BLAST search, the AB3 strain (producer strain) was 99% homologous to Bacillus malacitensis. Furthermore, the colonial morphology of the AB3 strain was applied to Bergey’s manual of determinative bacteriology, and the organism was identified as B. malacitensis, which confirms the identity of the bacteria.
Discussion
As the worldwide crisis of antibiotics resistance is still on-going, it has become obvious that seeking for new antibiotics and its alternative therapies is seriously essential.[1-33]Antibiotics alternative therapies (e.g., vaccines and AMPs) were claimed as promising candidate to replace traditional antibiotics or to be used in combination with clinically used antibiotics.[32-35] Thus, the aim of this research was to identify a novel AMP (bacteriocin) with potential activity against drug resistant pathogenic bacteria. The isolation, partial purification, and characterization of a potentially novel antimicrobial compound (named AB3), which acts against MRSA, MSSA, and B. subtilis, was investigated.
Although a total of eighty different bacterial strains/species were obtained from tested environmental samples, this would not reflect 100% recovery. This is because all used laboratory growth conditions might be not ideal for the complete recovery. In addition, about 5% of all screened bacterial isolates showed in vivo antagonistic activity towards tested indicators. This data should demonstrate the limited probability of obtaining antimicrobial producing strains. Nevertheless, it can be suggested that soil and water could be rich sources to screen for antimicrobial producing bacteria. In the simultaneous antagonism test, several not-medically important indicator strains were used, because these indicators may give a remarkable indication for the activity of the isolated peptide when certain pathogenic strains are not allowed to be examined. For instance, Bacillus anthracis which is a pathogenic bacterial strain that was not allowed to be tested in the used laboratory, thus the susceptibility of B. subtilis towards the isolated peptide was tested as it might be similar to other Bacillus species, including B. anthracis. In this experiment, susceptibility of B. subtilis to the isolated AB3 peptide was confirmed. However, this was not the case when B. cereus was tested against the isolated AB3 peptide, which was fully resistant.
Laboratory production of the peptide is very essential for its identity and characterization. The successful growth of the peptide producing strain in a broth medium does not necessarily means the targeted compound was produced/secreted.[36]This is why well diffusion assays were performed on supernatants of all tested broth media against B. subtilis (sensitive strain). The medium made of TSB without additives has led to the largest and clearest inhibitory zone (9 mm), while a smaller inhibitory zone (3 mm) was observed for TSB plus 5% sucrose. In addition, no antimicrobial activity (0 mm) was monitored for bacterial supernatants after growing the producing strain in TSB plus 5% lactose or TSB plus 5% glucose. It was claimed that the concentration and type of the used carbon source in a broth medium would influence the overall production of certain AMPs.[37]
Purification steps were successfully accomplished as only one fraction was active from each step of purification. This would indicate a successful binding of the AB3 peptide to the used column, thus separation from impurities was achieved. In addition, the size of inhibitory zone caused by the supernatant of AB3 strain was increased from 9 mm to 11 mm and from 11 mm to 14 mm following Strata and Sep-Pack chromatography, respectively. These increases in the potency (zone size) of the AB3 peptide towards the indicator strain (B. subtilis) following purification steps would also indicated a succeed purification for the AB3 peptide.
Klaenhammer has stated that almost 99% of all Gram-positive and Gram-negative bacteria presented in different environments are capable of producing at least one bacteriocin.[38] The analysis of 16SrRNA gene sequences and the colonial morphology of the AB3 producing strain has identified this bacterium as B. malacitensis, which was recovered from an environmental soil sample. Literatures have reported that the majority of antimicrobial Bacillus producing stains are isolated from soil samples.[39,40] Nevertheless, Bacillus species, which are Gram-positive, spore forming and aerobic bacteria, can be presented in different environments, including soil, dust, rocks, clays, and the gastrointestinal tracts of certain animals and insects.[41] The ability of this bacterium to survive in these diverse habitants might be linked with its spore formation that enables the genus of Bacillus to endure any harsh conditions, such as heat and dryness in various environments.[42]
MALDI-TOF analysis of the AB3 active fraction showed five peaks with the following molecular mass: 1288.207 Da, 1304.536 Da, 1326.529 Da, 1403.591 Da, and 1472.792 Da. In addition, AB3 peptide, that was produced by a Gram-positive strain (B. malacitensis), was revealed to be stable to high temperature when it was incubated at 75°C for 8 h during methanol evaporation. These findings might lead to the suggestion that the isolated AMP (AB3) is a member of lantibiotics (Class I). This is because class I of bacteriocins includes thermostable peptides with molecular mass <5 kDa that are produced by Gram-positive producing strains.[43]
Bacteriocin produced by Gram-positive producing strains are usually active against Gram-positive bacteria; however, they rarely act against Gram-negative bacteria.[44] Spectrum of activity of the isolated peptide was monitored against methicillin-resistant and methicillin-sensitive S. aureus (MRSA, MSSA)and B. subtilis, which are Gram-positive strains, and no activity was observed towards tested Gram-negative strains. This may support the suggestion that the isolated bacteriocin would possess a narrow spectrum of activity towards certain Gram-positive bacteria. Although the MIC of the AB3 peptide toward sensitive strains was determined, the exact MIC of the peptide for a potential use in human subjects would not be determined as the concentration of the used peptide was not quantified. Nevertheless, literatures have defined bactericidal agents as those with MBC/MIC ratios of ≤4.[45] The MBC/MIC ratios of the AB3 isolated bacteriocin against all tested strains were about 1, and this may indicate its bactericidal effect.
Although the in vivo activity of the isolated AB3 peptide against MSSA and MRSA (human pathogenic bacteria) is promising, further investigations to ensure its in vivo activity, spectrum of activity, safety, stability, and mode of action is essential to determine its suitability for potential applications, including potential therapeutic agent in human health. In addition, evaluation for any possible synergistic effect between AB3 and clinically used antibiotics could potentially lead to new combination therapy. Combination therapy increases the antimicrobial activity of both agents, reduces selection pressure of antibiotics and expands the antimicrobial spectrum. Increased antimicrobial activity due to a combination therapy reduces the MIC that is required for treating serious infections, such as those caused by multi-drug resistant organisms (MDROs).[46] Moreover, transposon mutagenesis is a technique that can be used to identify the gene responsible for the production of the AMP in the producing strain or whole genome sequencing to identify whole gene cluster encoding the compound. Once the responsible gene has been identified, synthetic production and protein modifications to improve the spectrum of activity or remove any toxic properties, if present, might become possible.
Conclusion
A novel bacteriocin with bactericidal effect toward MRSA, MSSA, and B. subtilis was isolated from B. malacitensis, which was recovered from an environmental soil sample. This isolated peptide belongs to Class I bacteriocin with molecular mass <5 kDa and stability to high temperature. This compound can potentially be used to treat serious infections caused by MSSA and MRSA or other infections caused by pathogenic Bacillus species. Nevertheless, more studies to ensure its safety and suitability for target application must be conducted.
Authors Declaration Statements
Ethics approval and consent to participate
This type of study is a non-human subject research and there was not any kind of individual participation, so the ethical approval and consent is exempt.
Availability of data and materials
The data used in this study are available and will be provided by the corresponding author on a reasonable request.
Competing interests
The author declared no conflict of interests.
Funding statement
This study is self-funded.
Author contribution
A.A designed the study, analyzed the data, interpreted results, and prepared the manuscript.
References
- Antimicrobial peptides against bacterial pathogens:Innovative delivery nanosystems for pharmaceutical applications. Antibiotics. 2023;12:184.
- [Google Scholar]
- 2018. Antibiotic Antimicrobial Resistance (AR/AMR):Centers for Disease Control and Prevention. Available from: https://www.cdc.gov/drugresistance/about.html
- Antibacterial drugs:Discovering antibiotics through soil metagenomics. Nat Rev Drug Discov. 2018;17:240-1.
- [Google Scholar]
- Something old, something new:Revisiting natural products in antibiotic drug discovery. Can J Microbiol. 2014;60:147-54.
- [Google Scholar]
- Antibiotic Resistance Threats in the United States 2013. Atlanta, Georgia: Centers for Disease Control and Prevention; 2013.
- 2018. Antimicrobial Resistance. Geneva: World Health Organization; Available from: https://www.who.int/news-room/fact-sheets/detail/antimicrobial-resistance
- Antibiotic resistance-the need for global solutions. Lancet Infect Dis. 2013;13:1057-98.
- [Google Scholar]
- 2016. Tackling Drug-resistant Infections Globally:Final Report and Recommendations:Review on Antimicrobial Resistance. Available from: https://amr-review.org/publications.html
- 2016. Infection Prevention, Control and Surveillance:Limiting the Development and Spread of Drug Resistance:Review on Antimicrobial Resistance. Available from: https://amr-review.org/publications.html
- Novel biotechnological applications of bacteriocins:A review. Food Control. 2013;32:134-42.
- [Google Scholar]
- Novel antimicrobial agents against multi-drug-resistant gram-negative bacteria:An overview. Recent Pat Antiinfect Drug Discov. 2012;7:175-81.
- [Google Scholar]
- Antimicrobial peptides produced by microorganisms. In: In:Antimicrobial Peptides and Innate Immunity. New York: Springer; 2013. p. :53-95.
- [Google Scholar]
- Bacteriocin gene-trait matching across the complete lactobacillus pan-genome. Sci Rep. 2017;7:3481.
- [Google Scholar]
- Discovery of a widely distributed toxin biosynthetic gene cluster. Proc Natl Acad U S A. 2008;105:5879-84.
- [Google Scholar]
- Classification of bacteriocins from gram-positive bacteria. In: In:Prokaryotic Antimicrobial Peptides. New York: Springer; 2011. p. :29-53.
- [Google Scholar]
- Bacteriocins-a viable alternative to antibiotics? Nat Rev Microbiol. 2013;11:95-105.
- [Google Scholar]
- A review of antimicrobial peptides and their therapeutic potential as anti-infective drugs. Curr Eye Res. 2005;30:505-15.
- [Google Scholar]
- Bacteriocins:Safe, natural antimicrobials for food preservation. Int J Food Microbiol. 2001;71:1-20.
- [Google Scholar]
- Different strategies for purification of antimicrobial peptides from lactic acid bacteria (LAB) In: Méndez-Vilas A, ed. Communicating Current Research Educational Topics Trends in Applied Microbiology.. Vol Vol. 2. Badajoz: Formatex; 2007. p. :557-68.
- [Google Scholar]
- Bacteriocins:Developing innate immunity for food. Nat Rev Microbiol. 2005;3:777-88.
- [Google Scholar]
- Lantibiotics:Peptides of diverse structure and function. Annu Rev Microbiol. 2007;61:477-501.
- [Google Scholar]
- Genetics of bacteriocins produced by lactic acid bacteria. FEMS Microbiol Rev. 1993;12:39-85.
- [Google Scholar]
- Antibacterial activities of bacteriocins:Application in foods and pharmaceuticals. Front Microbiol. 2014;5:241.
- [Google Scholar]
- Purification and characterization of nisin P produced by a strain of Streptococcus gallolyticus . J Med Microbiol. 2020;69:605-16.
- [Google Scholar]
- “Fingerprinting”beta-haemolytic streptococci by their production of and sensitivity to bacteriocine-like inhibitors. J Med Microbiol. 1979;12:397-411.
- [Google Scholar]
- BSAC Methods for Antimicrobial Susceptibility Testing, Version. Birmingham:British Society for Antimicrobial Chemotherapy 2013:1-87.
- [Google Scholar]
- BACTIBASE:A new web-accessible database for bacteriocin characterization. BMC Microbiol. 2007;7:89.
- [Google Scholar]
- BACTIBASE second release:A database and tool platform for bacteriocin characterization. BMC Microbiol. 2010;10:22.
- [Google Scholar]
- Alternatives therapeutic approaches to conventional antibiotics:Advantages, limitations and potential application in medicine. Antibiotics (Basel). 2022;11:1826.
- [Google Scholar]
- Increasing antimicrobial resistance poses global threat, WHO Says. JAMA. 2023;329:200.
- [Google Scholar]
- Futuristic non-antibiotic therapies to combat antibiotic resistance:A review. Front Microbiol. 2021;12:609459.
- [Google Scholar]
- Antimicrobial peptides:A potent alternative to antibiotics. Antibiotics (Basel). 2021;10:1095.
- [Google Scholar]
- Influence of media and temperature on bacteriocin production by Bacillus cereus 8A during batch cultivation. Appl Microbiol Biotechnol. 2004;65:158-62.
- [Google Scholar]
- Cultural conditions and nutritional components affecting the growth and bacteriocin production of Lactobacillus plantarum KC21. Food Sci Biotechnol. 2010;19:793-802.
- [Google Scholar]
- Antimicrobial producing microbes isolated from soil samples collected from Nanga Merit Forest in Sarawak, Malaysian Borneo. Eur J Exp Biol. 2014;4:494-501.
- [Google Scholar]
- Molecular characterization of bacterial isolates from soil samples and evaluation of their antibacterial potential against MDRS. Molecules. 2022;27:6281.
- [Google Scholar]
- A brief history of antibiotics and select advances in their synthesis. J Antibiot (Tokyo). 2017;71:153-84.
- [Google Scholar]
- Diversity and applications of Bacillus bacteriocins. FEMS Microbiol Rev. 2011;35:201-32.
- [Google Scholar]
- Bacteriocins and its use for multidrug-resistant bacteria control. Antibiot Resist. ;2016:329-49.
- [Google Scholar]
- Bacteriocins, antimicrobial peptides from bacterial origin:Overview of their biology and their impact against multidrug-resistant bacteria. Microorganisms. 2020;8:639.
- [Google Scholar]
- Characterization of variables that may influence ozenoxacin in susceptibility testing, including MIC and MBC values. Diagn Microbiol Infect Dis. 2014;78:263-7.
- [Google Scholar]
- Combinatory therapy antimicrobial peptide-antibiotic to minimize the ongoing rise of resistance. Front Microbiol. 2019;10:1703.
- [Google Scholar]







