Translate this page into:
Investigating the role of Vitamin D and its targeted gene-pathway interactions involved in cardiometabolic health
*Corresponding authors: Fazli Rabbi Awan, Health Biotechnology Division, National Institute for Biotechnology and Genetic Engineering, Faisalabad, Pakistan. awan.fr@gmail.com
Susan L. M. Coort, Department of BioinformaticsBiGCaT, NUTRIM School of Nutrition and Translational Research in Metabolism, Maastricht University, Maastricht, The Netherlands. susan.coort@gmail.com; susan.coort@maastrichtuniversity.nl
-
Received: ,
Accepted: ,
How to cite this article: Fiaz H, Khan AR, Ehrhart F, Hussain M, Evelo CT, Awan FR, et al. Investigating the role of Vitamin D and its targeted gene-pathway interactions involved in cardiometabolic health. Inter J Health Sci. 2025;19:29-42. doi: 10.25259/OA04_8774
Abstract
Objectives
Low levels of Vitamin D and its related gene variants are implicated in cardio-metabolic disorders (CMDs). This study aimed to elucidate the effect of Vitamin D deficiency and the relationship of genetic polymorphisms of Vitamin D synthesizing enzymes and Vitamin D receptor (VDR) with cardiovascular diseases (CVD) and comorbid conditions (hypertension [HTN] and diabetes mellitus [DM]).
Methods
This is a case-control study with a random sampling technique. Patients (n = 400) were having CVD (without any complication) and CVD (with HTN and/or DM). Healthy controls (n = 226) were without any disease. Vitamin D metabolites were measured in 30 controls and 51 CMD patients by liquid chromatography-mass spectrometry. Effect of five single-nucleotide polymorphisms (SNPs) of VDR (rs7975232 and rs2228570), CYP2R1 (rs10741657 and rs10766197), and CYP27B1 (rs10877012) on CMDs was tested. Furthermore, network analysis was performed to identify possible candidate genes and pathways linked to CMDs.
Results
CMD patients were Vitamin D deficient (Calcifediol, P = 0.006; Calcitriol, P = 0.005) relative to controls. Tested SNPs were found not to be associated with Vitamin D metabolites levels. Logistic regression models revealed heterozygous genotypes of rs2228570 (odds ratio [OR]: 1.12, 95% confidence interval [CI]: 0.6-2.08, P = 0.02) and rs10766197 (OR: 1.8, 95% CI: 1.1-2.93, P = 0.01) in the manifestation of HTN and DM in cardiovascular patients, respectively. Network analysis showed an association of several genes (i.e., tumor necrosis factor, parathyroid hormone, and fibroblast growth factor 23) linked to Vitamin D pathways.
Conclusions
SNP association and exploration of Vitamin D-SNP-Disease-Gene-Pathway networks may help in the effective management and treatment strategies for CMDs through personalized medicine.
Keywords
Cardiometabolic diseases
CYP27B1
CYP2R1
Polymorphisms
Single-nucleotide polymorphisms
Vitamin D receptor
Vitamin D deficiency
INTRODUCTION
Cardiovascular diseases (CVDs), diabetes mellitus (DM), and hypertension [HTN] are heterogeneous noncommunicable disorders that are caused by excessive and unhealthy food intake, physical inactivity, and genetic predisposition. In addition to these factors, lack of public awareness, limited health facilities, and poverty contribute to the burden of these diseases in the low- to middle-income countries. Co-occurrence of CVDs and any of the metabolic conditions such as DM, HTN, obesity, and dyslipidemia are termed as cardio-metabolic disorders (CMDs).[1] Patients with CMDs are 2-4 times more likely to die due to coronary heart diseases. Globally, about 25% of the adults are suffering from CMDs[2] which are a major reason for mortality around the globe. The death rate due to CVD, HTN, and diabetes is 44%, 14% and 4%, respectively.[3,4] Apart from major dietary components (i.e., carbohydrates, fats, and proteins), trace elements, minerals, and vitamins also play significant role in maintaining optimum health and an imbalance in these can lead to several diseases including CMDs.
Vitamins are organic compounds required by the body in small amounts for the regulation of metabolism. These are important in supporting the physiological functions of cells, tissues, and organs. Vitamins are usually classified as water-soluble (B complexes and C) and fat-soluble (A, D, E, and K).[5] Deficiencies or excess of these vitamins cause adverse health consequences. Among fat-soluble vitamins, rickets in children and osteomalacia in adults characterize Vitamin D deficiency. Besides these, Vitamin D deficiency and/or polymorphisms of Vitamin D-related genes have also been linked to non-communicable diseases, including CVDs, HTN, and DM.[6-8] Of the myriad of physiological actions of Vitamin D, it helps to regulate renin-angiotensinaldosterone function and mediates the synthesis of nitric oxide (NO) synthase enzyme. NO synthase is involved in NO production, which causes vasodilation. Molecular mechanisms underlying the development of CMDs due to Vitamin D deficiency remain elusive. Furthermore, some previous studies have not shown consistent results regarding cardiometabolic effects of Vitamin D.[9,10]
Regarding the synthesis of Vitamin D in the body, on exposure to sunlight cholecalciferol is synthesized under the skin. Subsequently, calcifediol (25-hydroxyvitamin D3 [25(OH)D3]) and calcitriol (1,25-dihydroxyvitamin D3 [1,25(OH)2D3]) are produced from cholecalciferol by hepatic 25-hydroxylase and renal 1a-hydroxylase, respectively. These enzymes are the functional products of CYP2R1 and CYP27B1 genes.[11] Calcitriol binds to the Vitamin D receptor (VDR) that is encoded by the VDR. In turn, Vitamin D-VDR interacts with another member of the nuclear receptor family, termed as retinoid X receptor (RXR). The Vitamin D-VDR-RXR assembly recognizes the specific sequences on the DNA strand, called Vitamin D response elements (VDRE). This Vitamin D-VDR-RXR complex recruits further co-modulators to trigger various genomic and non-genomic processes.[12-14]
Several studies have been conducted to evaluate the impact of single-nucleotide polymorphisms (SNPs) in Vitamin D-related genes with CMDs.[15,16] Advanced computational approaches have also been used to unravel mutual interactions among Vitamin D genes and metabolic diseases retrieved from various biological databases. Similarly, attempts have also been made to assess the influence of Vitamin D metabolites on the functions of various genes and their related biological pathways, such as renin-angiotensin-aldosterone and insulin signaling.[17,18]
Problem statement and research gap
Despite the facts described above, the findings of previous studies are inconsistent and conflicting regarding genetic variants of Vitamin D-related genes and complex metabolic disorders. In Pakistan, no such comprehensive study has been conducted to analyze SNPs of Vitamin D-related genes with cardiometabolic conditions such as CVD, HTN, and DM so far. Therefore, the present study aimed to investigate the association between SNPs of VDR (rs7975232 and rs2228570), CYP2R1 (rs10741657 and rs10766197), and CYP27B1 (rs10877012) in cardiovascular patients with HTN and DM. In addition, a network analysis was performed to explore 25(OH)D3 and 1,25(OH)2D3-targeted genes’ interaction with various biological processes linked with HTN and DM.
MATERIALS AND METHODS
Study subjects
A total of 626 human adult subjects, including 226 healthy controls and 400 cardiovascular patients were recruited for this case-control study by employing a random sampling technique. According to the WHO sample size calculator, the calculated sample size was 381 by considering a confidence interval (CI) of 95%, a margin of error of 0.05, and a risk factor prevalence of 0.46. Due to convenient sampling, a slightly higher sample number (n = 626) was collected. Patients were sampled from the Allied Hospital and Faisalabad Institute of Cardiology, Faisalabad, Pakistan. Patients were suffering from cardiovascular disorders, type 2 diabetes, and HTN. Therefore, patients were termed as cardiometabolic disease (CMD) patients. Sampling of healthy control subjects (without any disease) was done from the same city. The clinical and biochemical parameters and the genotyping of five SNPs (rs7975232, rs2228570, rs10741657, rs10766197, and rs10877012) pertaining to Vitamin D-related genes were measured and identified in all study subjects, respectively. From these subjects, a subset of 81 (healthy = 30; cardiometabolic patients = 51) was selected for the measurement of Vitamin D3 metabolites (calcifediol and calcitriol) by liquid chromatography-tandem-mass-spectrometry (LC-MS/MS) at the Radboud University Medical Center, Nijmegen, Netherlands. Patients included in this subset were having CVD, HTN, and DM, concurrently.
Ethical approval
This study was approved by the Ethical Review Committee of the National Institute for Biotechnology and Genetic Engineering, Faisalabad, Pakistan. All the participants provided oral and/or written informed consent. Moreover, a questionnaire including anthropometric and demographic information and medical history was collected for all the study participants.
Biochemical measurements
Blood samples were taken in gel-containing vacutainers and were centrifuged for 5-7 min at 4000 rpm. The serum was separated and stored at −20°C. Blood glucose level, liver function tests (LFTs), and renal function tests (RFTs) readings were taken through a semi-automated clinical chemistry analyzer (Microlab-300, Merck, Germany).[19] Systolic and diastolic blood pressure (BP) were measured using a sphygmomanometer and BP value >130/80 mmHg was considered for HTN.[20,21] The subjects who were taking anti-diabetic drugs or had fasting blood glucose levels >126 mg/dL were considered diabetic patients. Serum Vitamin D metabolites were analyzed through liquid chromatography-tandem-mass-spectrometry. 25(OH)D3 and 1,25(OH)2D3 were measured based on the method described by Ter Horst et al.[22] and Dirks et al.,[23] respectively.
SNP genotyping
SNP genotyping was performed as reported in our previous study.[24] Briefly, for genetic analysis, whole blood samples were collected in ethylenediaminetetraacetic acid (EDTA)-coated vacutainers, which were placed and stored in the refrigerator at 4°C. These samples were processed to extract the genomic DNA using the organic method (Phenol-Chloroform method).[24] The extracted DNA samples were kept at −20°C until further analysis. All samples were genotyped for the VDR SNPs (rs7975232 [C/A], rs2228570 [A/G]), CYP2R1 SNPs (rs10741657 [A/G], rs10766197 [G/A]), and CYP27B1 SNP (rs10877012 [G/T]). To collect the genotyping data of the SNPs, tri and tetra Amplification Refractory Mutation System-Polymerase Chain Reaction (ARMS-PCR) assays were performed on T100TM thermal cycler (Bio-Rad Laboratories Inc., America). The primer sets used for tetra ARMS-PCR and tri ARMS-PCR were designed in freely available primer1 (http://primer1.soton.ac.uk/primer1.html) and primer3 software (http://bioinfo.ut.ee/primer3-0.4.0/). Primer sequences and PCR conditions are provided in Supplementary Tables 1S and 2S. PCR results were validated by Sanger DNA sequencing, as given in the supplementary file [Figure 1S].
Statistical analysis
Statistical analysis was performed by R programming v.4.1 using selected R packages (readxl v.1.3.1, fBasics v.3042.89.1, dplyr v.1.0.7, tidyverse v.1.3.1, rstatix v.0.7.0, ggpubr v.0.4.0, ggplot2 v.3.3.5, viridis v.0.6.1, and SNPassoc v.2.0.2). The continuous variables were expressed as mean ± standard deviation, while categorical variables were represented as frequencies (percentage). Non-parametric tests were applied due to the non-normal distribution of data. Wilcoxon rank-sum test/Mann-Whitney U-test was applied to analyze the differences in the biochemical and clinical parameters between healthy and cardiometabolic patients. The influence of various genotypes of selected polymorphisms on biochemical parameters was assessed following Kruskal-Wallis test with Pairwise Wilcoxon test. Furthermore, Chi-square statistics was applied to compare the genotypic frequencies of patient and healthy groups. In addition, it was hypothesized that the studied polymorphisms might be associated with the disease; therefore, multinomial logistic regression analysis was performed to check the risk assessment of cardio-metabolic diseases associated with the selected variants. P < 0.05 was taken as significant.
Bioinformatics analysis
Various online databases were searched to retrieve information regarding (i) measured SNPs related to Vitamin D metabolism, (ii) Vitamin D target genes and their interactions, (iii) cardiometabolic phenotypes such as diabetes and HTN, and (iv) disease and Vitamin D-related biological processes.
First, an in-depth description of the investigated SNPs (rs7975232, rs2228570, rs10741657, rs10766197, and rs10877012) was obtained using the Ensembl variant effect predictor (VEP).[25,26] VEP provides a detailed annotation of the SNPs, such as variant position on chromosome, regional location on DNA sequence (intronic, exonic, and regulatory region), and the fallouts caused by the variant (pathogenicity/non-pathogenicity). Details are given in Table 1.
| Category | Description | ||||
|---|---|---|---|---|---|
| Gene/Protein product/Location |
VDR/Vitamin D Receptor/ 12: 47,841,537-47,943,048 |
CYP2R1/25-hydroxylase/11: 14,877,440-14,892,231 | CYP27B1/1α-hydroxylase/12: 57,762,334-57,768,986 | ||
| Gene ID | ENSG00000111424 | ENSG00000186104 | ENSG00000111012 | ||
| SNP | rs7975232 | rs2228570 | rs10741657 | rs10766197 | rs10877012 |
| Location | 12:47845054 Reverse strand |
12:47879112 Reverse strand |
11:14893332 Forward strand |
11:14900334 Forward strand |
12:57768302 Reverse strand |
| Allele | (C/A) | (A/G) | (A/G) | (G/A) | (G/T) |
| Minor Allele Frequency | 0.52 | 0.67 | 0.69 | 0.33 | 0.35 |
| Consequence | Intron variant | Loss of start codon | Upstream gene variant | Upstream gene variant | Upstream gene variant |
| Impact | Modifier | High | Modifier | Modifier | Modifier |
| Biotype | Protein coding | Protein coding | Protein coding | Protein coding | Protein coding |
| Exon, Intron | 9/9I | 1/8E | ----- | ----- | ----- |
| Amino acid | ------ | M/T | ----- | ----- | ----- |
| Codons | ------ | ATG/ACG | ----- | ----- | ----- |
| Distance to transcript | ------ | ------ | 1101 | 4663 | 1224 |
| SIFT | ----- | 0 | ----- | ----- | ----- |
| PolyPhen | ----- | 0.99 | ----- | ----- | ----- |
| Clinical significance | Benign, Likely pathogenic | Benign, drug response | ----- | ----- | ----- |
| CADD | 0.15 | 3.21 | −0.17 | 0.29 | 0.14 |
| Associated phenotypes | Vitamin D dependent rickets IIA | Vitamin D dependent rickets type II, Eosinophil counts | Vitamin D insufficiency | ----- | Vitamin D hydroxylation-deficient rickets type 1a, Hypocalcemic Vitamin D dependent rickets |
| DisGeNET | ----- | CAD, DM | CHD, DM | Non-insulin dependent DM, Vitamin D deficiency | ----- |
The results retrieved from VEP Ensembl and dbSNP database. VEP: Variant effect predictor, VDR: Vitamin D receptor, SNP: Single-nucleotide polymorphism, CAD: Coronary artery disease, CHD: Coronary heart disease, DM: Diabetes mellitus, SIFT: Sorting intolerant from tolerant, CADD: Combined annotation dependent depletion
Second, biological networks were created to investigate the overlap between known Vitamin D target-related genes and disease-associated genes. Vitamin D metabolite-gene and disease-gene networks were constructed in Cytoscape v.3.10.0.[27] The target genes of both calcifediol and calcitriol were selected from the comparative toxicogenomics database (CTD).[28,29] Protein-protein interactions between proteins known to be associated with a specific disease were assessed by the search tool for the retrieval of interacting genes/proteins (STRING) disease query using the String app in the Cytoscape. Moreover, to analyze disease-genes association, the datasets for HTN and DM were retrieved from STRING database[27] and DisGeNET.[30] Both for calcifediol and calcitriol, the chemical-gene networks were separately merged with the disease-gene networks and then extended to the pathways using CyTargetlinker plugin[31] and WikiPathways linkset.[32] These extended Vitamin D-SNP-Gene-Pathways networks were manually analyzed and a subset of each network was created based on the genes (VDR, CYP2R1, and CYP27B1) considered in the present study. Moreover, the studied SNPs (rs7975232, rs2228570, rs10741657, rs10766197, and rs10877012) linked to the aforementioned Vitamin D-related genes were manually added to these created subnetworks. The experimental workflow is given in Figure 1.
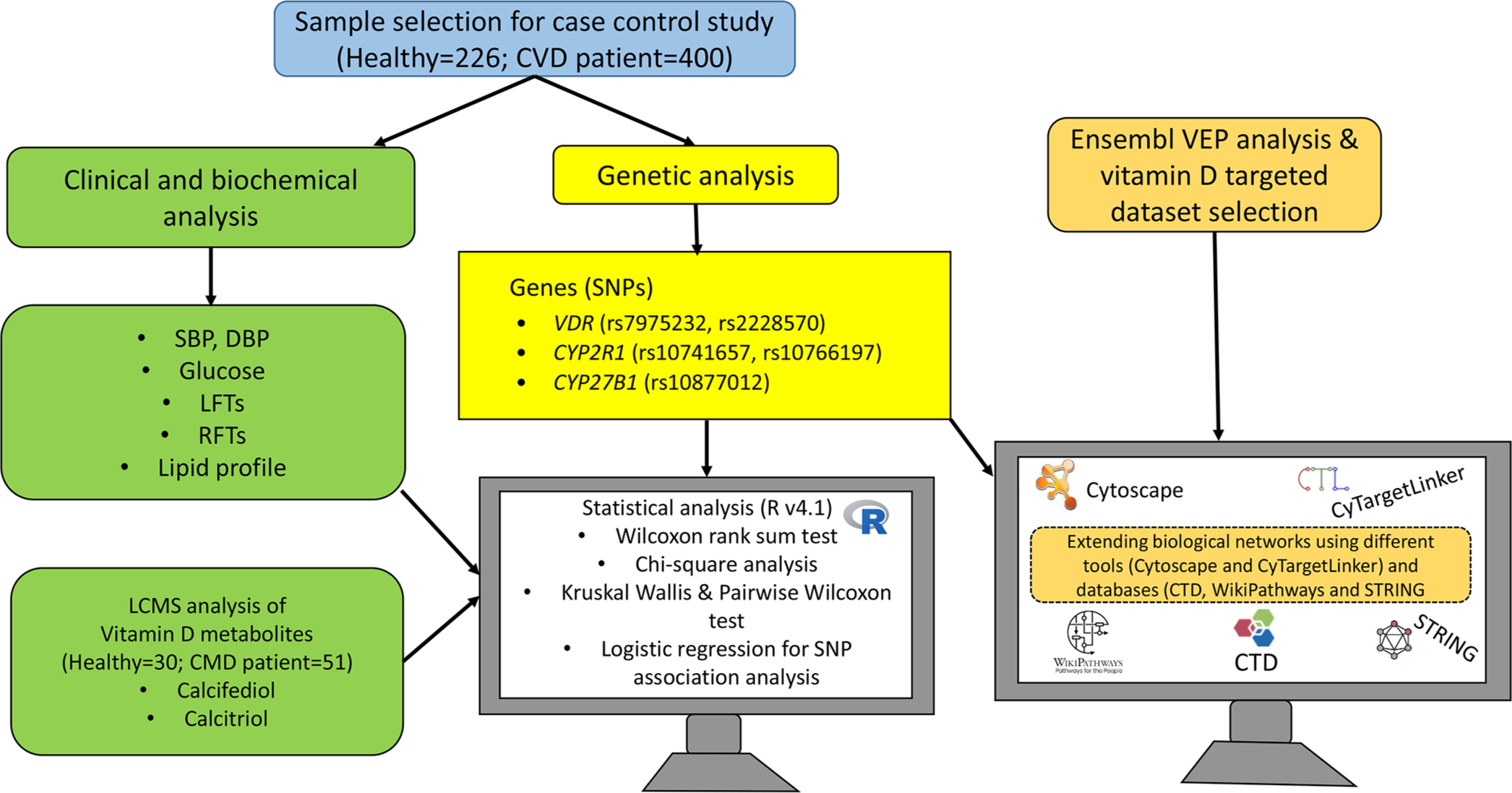
- Schematic representation of experimental and bioinformatics workflow. CVD: Cardiovascular disease, VEP: Variant effect predictor, VDR: Vitamin D receptor, SBP: Systolic blood pressure, DBP: Diastolic blood pressure, LFTs: Liver function test, RFTs: Renal function test, LCMS: Liquid chromatography-tandem mass spectrometry, CMD: Cardio-metabolic diseases, SNP: Single nucleotide polymorphisms, CTD: Comparative toxicogenomics database, STRING: Search tool for the retrieval of interacting genes/proteins.
RESULTS
For the evaluation of effects of genotypes from the studied SNPs on the clinical and biochemical parameters (i.e., systolic BP, diastolic BP, blood glucose level, RFTs, LFTs, lipid profile, and Vitamin D metabolites), Kruskal-Wallis and pairwise Wilcoxon tests were applied. For the control group, a significant difference was found for systolic BP (P = 0.003) and diastolic BP (P = 0.01) between GG and AG genotypes of rs2228570 polymorphism. Low-density lipoprotein cholesterol (LDL-C) was found to be significantly different between AA and CA (P = 0.04), and between AA and CC (P = 0.003) genotypes of rs7975232 polymorphism, given in Supplementary Figure 2SA and Table 3S.
Some of the biochemical parameters were found to be significantly different between different genotyping groups of CVD patients. Patients carrying AG or GG genotypes (rs2228570 A/G polymorphism) had significant differences in systolic BP (P = 0.009) and LDL-C (P = 0.0007). Patients having CA and CC genotypes (rs7975232 C/A polymorphism) were found to be different for LDL-C (P = 0.03) and high-density lipoprotein cholesterol (P = 0.001), as given in Supplementary Figure 2SB and Table 3S. The biochemical parameters were not statistically different for genotypes of polymorphisms (rs10741657, rs10766197, and rs10877012) for CYP2R1 and CYP27B1 genes given in Supplementary Figures 3S and 4S. Similarly, calcifediol and calcitriol did not show a significant association with any of the abovementioned polymorphisms.
Furthermore, the association of SNPs with diseases was analyzed using logistic regression. This revealed that polymorphisms, that is, rs2228570 A/G and rs10766197 G/A were associated with HTN and DM in CMD patients. The codominant model showed that AG genotype of rs2228570 is associated with a higher risk of HTN (odds ratio [OR]: 1.12, 95% CI: 0.6-2.08, P = 0.02). From the recessive model, it was also inferred that the homozygous major genotype (AA) of rs2228570 played a protective role from HTN in CMD patients (OR: 0.18, 95% CI: 0.06-0.59, P = 0.006). CMD patients seemed to be at higher risk (~80%) of diabetes development having rs10766197 GA genotype. This result was supported by both codominant (OR: 1.8, 95% CI: 1.1-2.93, P = 0.01) and over-dominant (OR: 1.89, 95% CI: 1.23-2.9, P = 0.004) models.
To assess the relationship of biochemical parameters from a disease perspective, a subset of 81 subjects were chosen, including healthy controls (n = 30) and CMD patients (n = 51). Wilcoxon rank sum Test/Mann-Whitney U-test was applied to determine the difference of various biochemical variables in CMD patients versus healthy controls. Results showed that BP, liver enzymes (i.e., alanine aminotransferase, aspartate aminotransferase, and alkaline phosphatase), glucose, uric acid, creatinine, and urea were found to be higher in patients than the healthy control subjects. On the other hand, calcifediol and calcitriol levels were significantly decreased in patients relative to the healthy controls, as given in Table 2.
| Variables | Controls (n=30) | Patients (n=51) | P-value | Ref range |
|---|---|---|---|---|
| Systolic BP (mmHg) | 120±04 | 144±26 | 2.6×10−6**** | ≤120 |
| Diastolic BP (mmHg) | 80±05 | 92±15 | 2.02×10−4*** | ≤80 |
| Glucose (mg/dL) | 91±21 | 216±105 | 3.82×10−9**** | Random: <200 Fasting: <126 |
| Uric acid (mg/dL) | 5.5±1.8 | 7.0±3 | 1.81×10−2* | 2.5-7.7 |
| ALT (U/L) | 19±09 | 24±16 | 1.56×10−1Ns | 9-43 |
| AST (U/L) | 21±08 | 28±17 | 5.09×10−2 Ns | 10-35 |
| ALP (U/L) | 153±50 | 207±63 | 1.90×10−4*** | 65-270 |
| Creatinine (mg/dL) | 0.7±0.2 | 1.3±0.7 | 2.29×10−4*** | 0.6-1.1 |
| Urea (mg/dL) | 22±7 | 51±33 | 1.88×10−6 **** | 10-50 |
| Albumin (g/dL) | 4.1±0.2 | 3.8±0.8 | 3.69××10−4*** | 3.5-5.0 |
| Cholesterol (mg/dL) | 171±34 | 160±40 | 2.41×10−1Ns | <200 |
| HDL-C (mg/dL) | 46±6 | 53±10 | 2.02×10−3** | >35 |
| LDL-C (mg/dL) | 83±8 | 71±10 | 5.63×10−6**** | <130 |
| Triglycerides (mg/dL) | 239±145 | 227±111 | 4.84×10−1Ns | <150 |
| Calcifediol (nmol/L) | 44±25 | 31±21 | 0.006** | >75 |
| Calcitriol (pmol/L) | 145±33 | 83±45 | 0.005** | >187 |
This tableincludes the result for subset cohort whose Vitamin D metabolites were measured. BP: Blood pressure, ALT: Alanine aminotransferase, AST: Aspartate aminotransferase, ALP: Alkaline phosphatase, HDL-C: High-density lipoprotein cholesterol, LDL-C: Low-density lipoprotein cholesterol. Ns, *, **, *** and **** show P≥0.05, < 0.05, <0.01, <0.001, and<0.0001
Furthermore, five studied SNPs were subjected to VEP analysis in the Ensembl. Several properties such as minor allele frequency, consequence, impact, biotype, codon change, sorting intolerant from tolerant (SIFT), PolyPhen, and combined annotation dependent depletion (CADD) scores related to the impact of variants on protein function were collected from Ensembl VEP analysis, and the dbSNP database is given in Table 1. Among these, SIFT (0-0.05; deleterious and 1.0; tolerated), PolyPhen (0-0.15; benign and 0.15-1.0; damaging), and CADD (cutoff score is 20; below this value, the variant is considered as benign and above 20 the variants are classified as harmful) are pathogenicity prediction tools/scores. Moreover, VEP analysis also provides information about the association of variants with phenotypes by retrieving information from one of the largest databases, such as DisGeNET.[33]
Network analysis
Inter-relationship of Vitamin D metabolites, HTN, and diabetes-related genes
First, Vitamin D-SNP-disease-protein pathway networks were generated to explore biological pathways connected with VDR, CYP2R1, CYP27B1, and Vitamin D3-related SNPs rs7975232, rs2228570, rs10741657, rs10766197, and rs10877012. Furthermore, these proteins and biological pathways linked with Vitamin D metabolites were also identified and investigated for disease conditions such as CVD, HTN, and DM.
The CTD and STRING databases were used to retrieve biochemical-protein interactions and protein-protein interactions, respectively, to create networks for the identification of proteins targeted by 25(OH)D3 and 1,25(OH)2D3 and associated with HTN and DM. For disease query in STRING database, confidence score of 0.9 was used. The Vitamin D metabolites targets from CTD and the protein-protein interactions from STRING were merged and four networks were generated depending on the Vitamin D metabolite and disease of interest. Each network was extended with protein-pathway interactions from WikiPathways. Vitamin D-disease-protein pathway networks were built to explore the biological pathways that were directly linked to the genes (VDR, CYP2R1, and CYP27B1) under investigation in the present study [Figures 2-5]. Table 3 enlisted the pathways represented in the created disease-Vitamin D metabolite networks.
| Pathway name | 25(OH) D3 target genes | 1,25(OH) 2D3 target genes | Hypertension-linked genes | Diabetes-linked genes | Pathway ID |
|---|---|---|---|---|---|
| Drug induction of bile acid pathway | 02 | 02 | 01 | 0 | https://www.wikipathways.org/instance/WP2289 |
| Meta-pathway biotransformation phase I and II | 07 | 04 | 08 | 0 | https://www.wikipathways.org/instance/WP702 |
| Non-genomic actions of 1,25 dihydroxyvitamin D3 | 04 | 10 | 07 | 05 | https://www.wikipathways.org/instance/WP4341 |
| Nuclear receptors | 01 | 03 | 04 | 03 | https://www.wikipathways.org/instance/WP170 |
| Nuclear receptors in lipid metabolism and toxicity | 04 | 05 | 06 | 02 | https://www.wikipathways.org/instance/WP299 |
| Nuclear receptors meta-pathway | 06 | 15 | 23 | 20 | https://www.wikipathways.org/instance/WP2882 |
| Ovarian infertility genes | 01 | 01 | 00 | 02 | https://www.wikipathways.org/instance/WP34 |
| Oxidation by cytochrome P450 | 07 | 04 | 08 | 0 | https://www.wikipathways.org/instance/WP43 |
| PI3K/AKT/mTOR -VitD3 Signaling | 02 | 03 | 03 | 04 | https://www.wikipathways.org/instance/WP4141 |
| Vitamin D in inflammatory diseases | 03 | 03 | 03 | 01 | https://www.wikipathways.org/instance/WP4482 |
| Vitamin D metabolism | 06 | 03 | 01 | 0 | https://www.wikipathways.org/instance/WP1531 |
| Vitamin D receptor pathway | 11 | 13 | 15 | 13 | https://www.wikipathways.org/instance/WP2877 |
| Vitamins A and D -action mechanisms | 01 | 01 | 0 | 0 | https://www.wikipathways.org/instance/WP4342 |
The table includes pathways, the total number of genes targeted by the Vitamin D3 metabolites (25(OH) D3 and 1,25(OH) 2D3) or linked to hypertension and diabetes. To view the pathway in WikiPathways, respective link is provided in the last column (Pathway ID). 25(OH) D3: 25-hydroxyvitamin D3. PI3K/AKT/mTOR: Phosphatidylinositol 3-kinase/Protein kinase B/Mammalian target of rapamycin
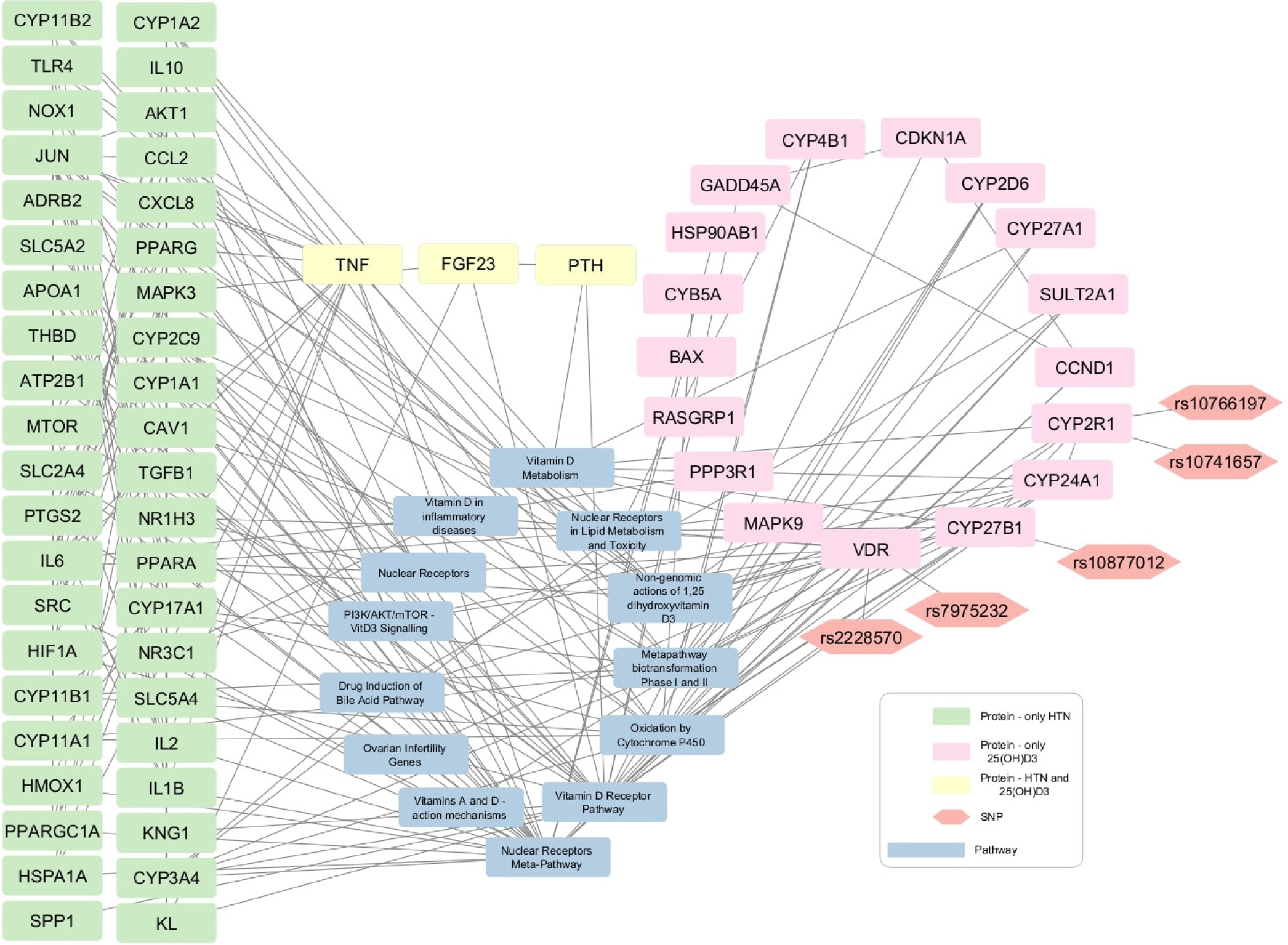
- 25-hydroxyvitamin D3 (25(OH)D3)-Hypertension network contains 42 protein nodes that are associated with hypertension (green rectangular shapes), 17 protein nodes are related with 25(OH)D3 (pink rectangular shapes), and three nodes are commonly linked to both hypertension and 25(OH)2D3 (yellow rectangular shapes). The gray rectangles show pathways and the orange hexagons represent the SNPs.
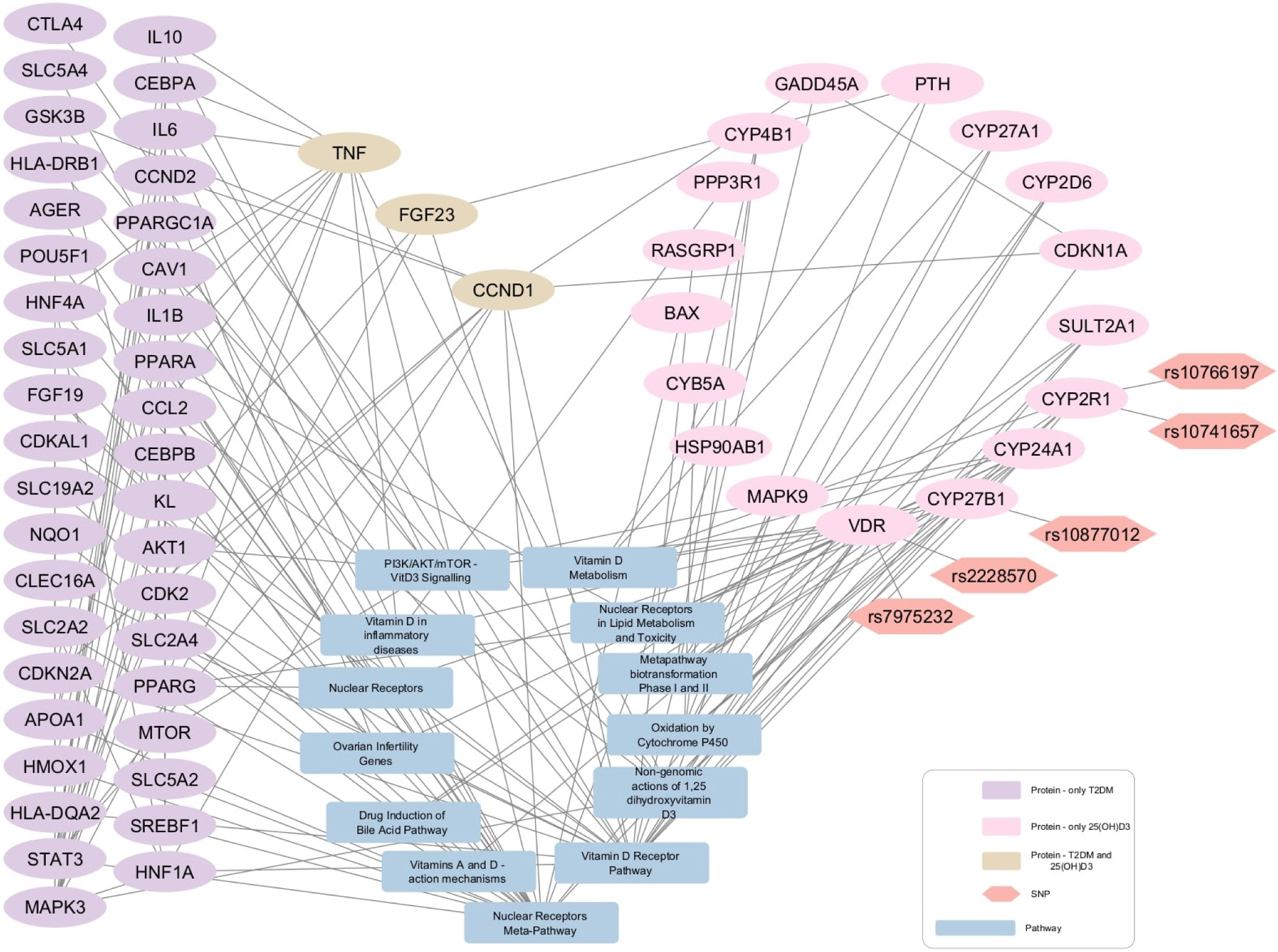
- Protein-pathway interaction network for diabetes mellitus and 25-hydroxyvitamin D3 (25(OH)D3). 25(OH)D3-Diabetes network 39 protein nodes are associated with diabetes (purple elliptical shapes), 17 protein nodes are related with 25(OH)D3 (pink elliptical shapes), and three nodes are commonly linked to both diabetes and 25(OH)D3 (Beige elliptical shapes). The gray rectangles show pathways and the orange hexagons represent the SNPs.
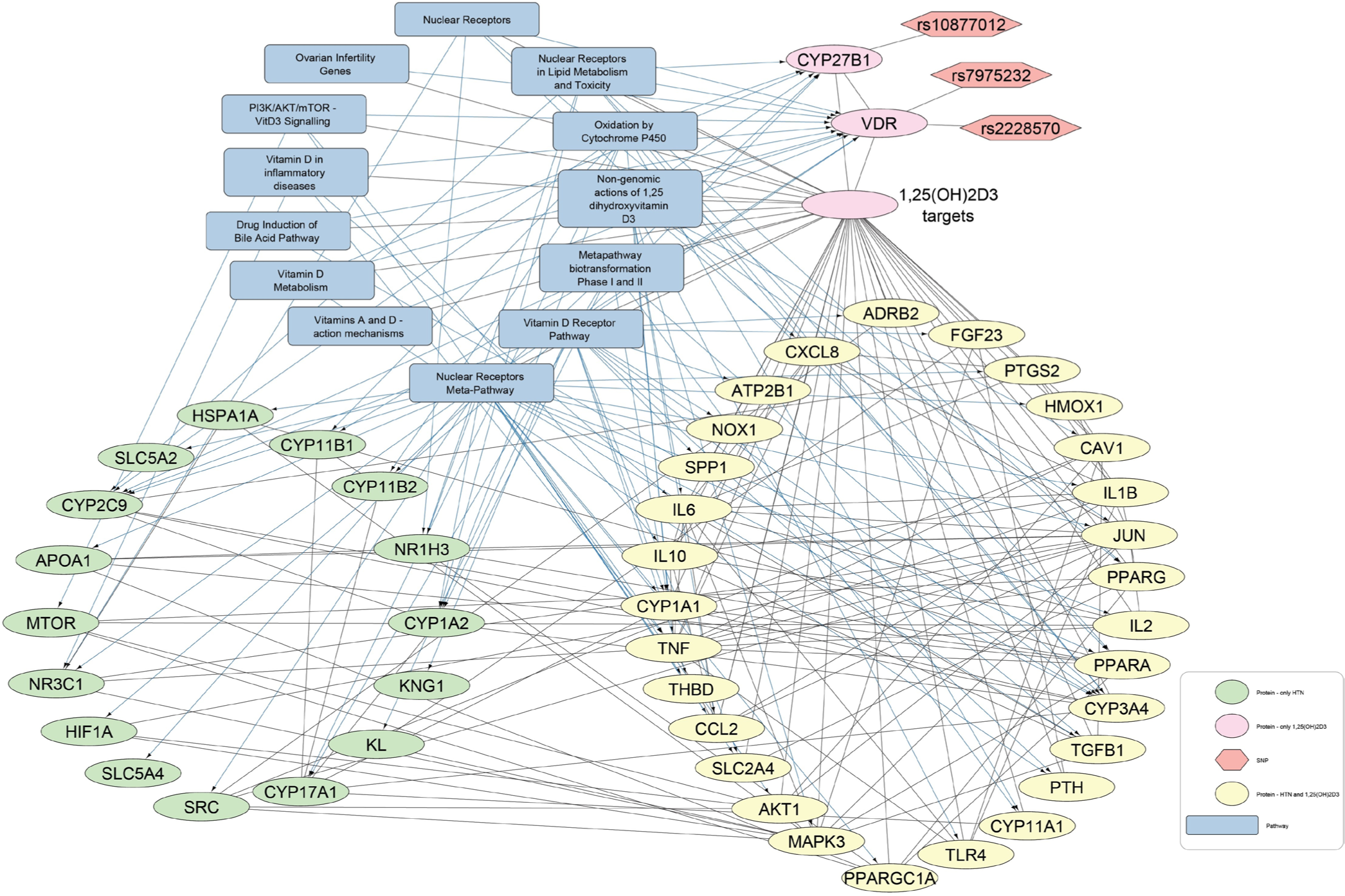
- Protein-pathway interaction network for hypertension and 1,25-dihydroxyvitamin D3 (1,25(OH)2D3). In this network of 1,25(OH)2D3-Hypertension, 16 protein nodes are associated with hypertension (green elliptical shapes), two protein nodes are related with 1,25(OH)2D3 (pink elliptical shapes), and 29 nodes are commonly linked to both hypertension and 1,25(OH)2D3 (yellow elliptical shapes). The gray rectangles show pathways and the orange hexagons represent the SNPs.
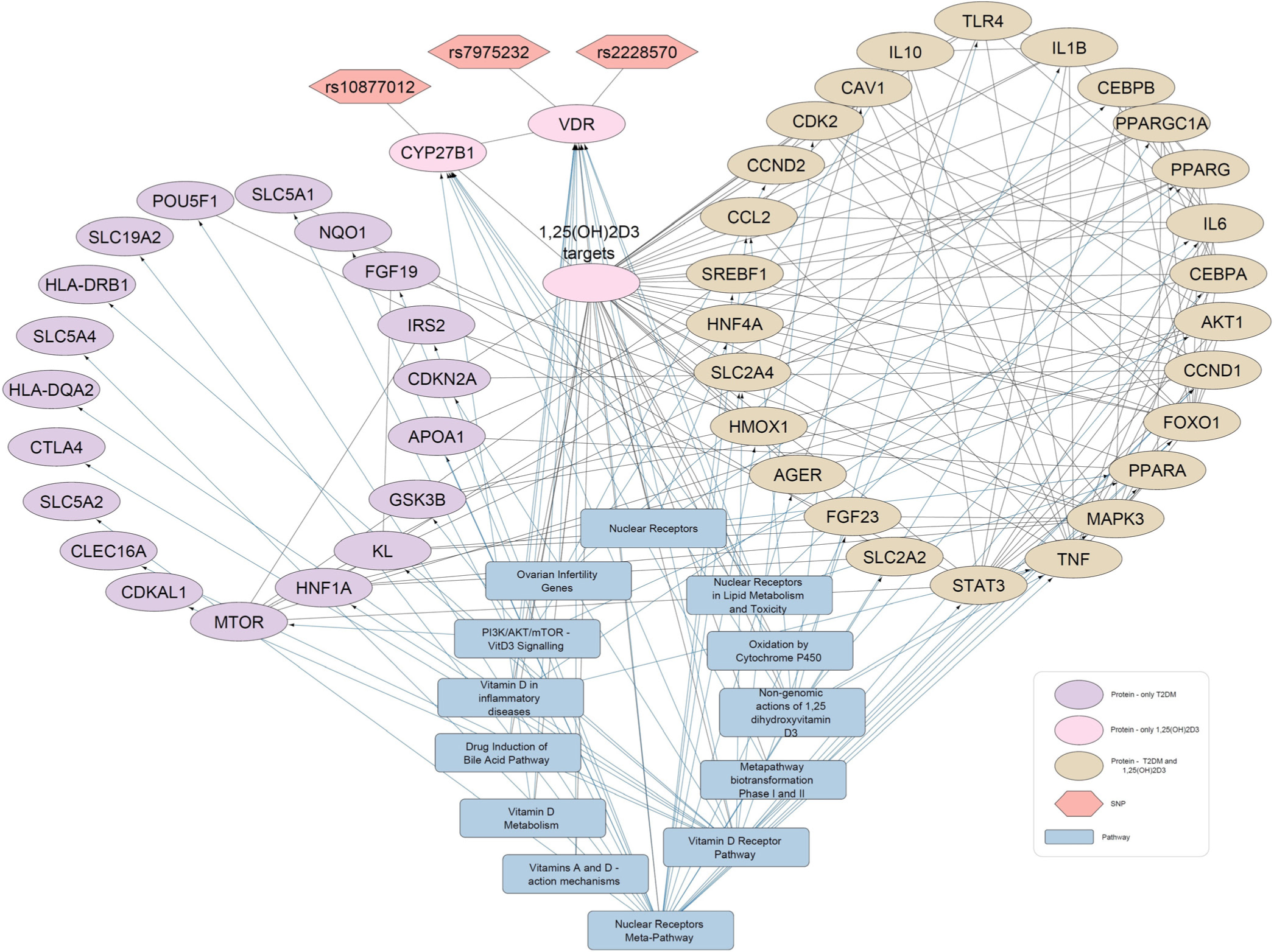
- Protein-pathway interaction network for diabetes mellitus and 1,25-dihydroxyvitamin D3 (1,25(OH)2D3). In the network of 1,25(OH)2D3-Diabetes, 26 proteins (Beige elliptical shapes) are commonly interacted with both 1,25(OH)2D3 and diabetes mellitus. The gray rectangles show pathways and the orange hexagons represent the SNPs.
In the four Vitamin D-disease-protein pathway networks [Figures 2-5], 13 biological pathways were similar. However, in each network, these pathways link with genes/proteins involving HTN and diabetes with varying degrees of interactions as the number of edges by which each pathway is linked with different genes is different.
Network analyses manifested that several proteins related to HTN and diabetes appeared to be present in Vitamin D3-related pathways. This suggests that Vitamin D3 may be involved in the modulation of various genes associated with HTN and diabetes. Some of the pathways and proteins that are shared by Vitamin D3 metabolites, HTN, and diabetes are discussed below.
First the VDR and nuclear receptor pathways are activated by binding of 1,25(OH)2D3. Successively, these activated pathways interact with various proteins that lead to downstream regulation of HTN- and DM-related genes, including tumor necrosis factor (TNF), parathyroid hormone (PTH), fibroblast growth factor 23 (FGF23), solute carrier family 2 member 4 (SLC2A4), and klotho protein (KL). VDREs, transcription factor VDR, and nuclear receptors might be contributing to the regulation of these genes. Moreover, it is shown that TNF interacts with proteins involved in blood glucose regulation and HTN. Moreover, TNF is also present in the Vitamin D-related inflammatory disease pathway, nuclear receptor meta-pathway, and nongenomic action of 1,25(OH)2D3. Overall, these Vitamin D-linked pathways regulate serum levels of TNF.
Interestingly, a recurrent protein FGF23 in the disease-Vitamin D metabolite networks has also been reported as a positive modulator of the renin-angiotensin system that underlies HTN pathogenesis.[34] FGF23 association with mitogen-activated protein kinase 3 (MAPK3) and KL (common components of the study networks) has also been manifested earlier that indicates their role in HTN and diabetes development.[35,36]
In addition to this, PTH is found to be involved in the regulation of HTN [Figure 2 and 4]. It is present in the VDR pathway and Vitamin D metabolism pathway. In addition, kininogen-1, which is an important protein of blood coagulation, is present in the VDR pathway [Figures 2 and 4]. This protein also shows an association with the renin and ACE inhibitor pathway involved in BP regulation. A protein, Cyclin D1 (CCND1), is seen to interact with 25(OH)D3 and diabetes [Figure 3]. The same gene is further linked with many proteins underlying diabetes pathogenesis. CCND1 is connected to the nuclear receptor and VDR pathways that might regulate CCND1 expression.
Furthermore, SLC2A4 appeared to be a target of 1,25(OH)2D3 and is associated with HTN and diabetes. Studies demonstrated that Vitamin D3 up-regulates the expression of glucose transporter 4 (GLUT4) protein channel (encoded by SLC2A4). GLUT4 is also known as solute carrier family 2 member 4, involving in glucose uptake by body cells keeping its level within permissible limits.[37,38]
DISCUSSION
Vitamin D is an essential nutrient for optimum health. It regulates multiple physiological functions such as mineralization, cellular growth, DNA repair, cell differentiation, apoptosis, membrane transport, cellular metabolism, and oxidative stress.[39,40] On the other hand, its deficiency may cause several health consequences, including osteoporosis, rickets, immune disorders, cancer, cardiovascular, and metabolic diseases. The present study aimed to test the association of serum Vitamin D levels, selected Vitamin D-related polymorphisms, and biochemical parameters with CVD, HTN, and diabetes.
Statistical analysis revealed that VDR SNPs (rs7975232 and rs2228570) were found to be associated with systolic BP, diastolic BP, low-density lipoprotein-cholesterol, and high-density lipoprotein-cholesterol. Pertinent to health conditions, rs2228570 and rs10766197 were found to be associated with HTN and DM in cardiac patients, respectively. The correlation between serum Vitamin D level and its related SNPs was found to be non-significant. This indicates Vitamin D level and its SNPs may link with cardiometabolic disorders, independently. A recent review mentioned the association of low Vitamin D levels with cardiovascular-related morbidity, and the haplotype analysis predicted the marginal association of study SNPs with lower risk of high BP.[41]
So far, various SNPs of VDR, CYP2R1, and CYP27B1 have been analyzed. Scazzone et al. reported a linear relationship of rs10766197 (CYP2R1) with metabolic syndrome. In this study, AA genotypic and allelic frequencies were found to be higher in metabolic syndrome patients relative to their healthy controls.[42] Another study manifested that GG genotype of rs12794714 and AA genotype of rs10766197 showed significant association with high risk of developing type 1 diabetes.[43] A case-control study revealed rs10741657 polymorphism as a novel single-nucleotide variant contributing to coronary artery disease.[44] Wang et al.[45] indicated AG and GG genotype of rs10766197 association with type 2 diabetes, but SNPs and serum Vitamin D status were not found to be correlated. The finding of our study about genotypes (AG and GG), SNPs, and serum Vitamin D levels are in accordance with the results reported by Wang et al.[45] According to Hussein et al., GG genotype carriers of rs10766197 and CC genotype carriers of rs10877012 are 2.6 times and 3.7 times more prone to type 1 diabetes, respectively. In the case of GG and CC genotypes’ synergistic effect, the propensity to develop type 1 diabetes becomes more pronounced.[46] A case-control study demonstrated that rs2228570 is a risk factor for type 2 diabetes development in adults.[47] It is also reviewed that VDR polymorphisms might be associated with the onset of HTN.[48,49] A European cohort study has reported a significant relationship between rs1544410 (T/T) and rs731236 (G/G) genotype VDR polymorphism with coronary artery disease, but the association between rs2228570 with coronary artery disease was non-significant.[50] Meta-analyses documented the ApaI, FokI, and CYP2R1 polymorphisms as genetic susceptibility factors in the development of coronary artery diseases.[51,52] A research study reported a lower serum 25-hydroxyvitamin D level in metabolic syndrome patients than healthy controls. This study did not find any difference in frequency distribution for polymorphisms of Vitamin D binding proteins, while in the case of CYP27B1 polymorphisms, only wild-type genotype was found among healthy controls and metabolic syndrome patients.[53]
Regarding the association of Vitamin D with its related SNPs, Krasniqi et al. established an association of GC and CYP2R1 polymorphisms with serum Vitamin D levels.[54] Similarly, the relationship between FokI C allele carriers and reduced Vitamin D status has been reported.[55,56] However, these correlations are not compatible with the findings of the present study, which might be due to a small study population (n = 81) and ethnic differences. Thus, large-scale prospective studies are warranted to establish clinically meaningful results.
As in the present study, bioinformatics tools were also applied to analyze interactions of 25(OH)D3, 1,25(OH)2D3, HTN, and DM with proteins and biological pathways, which were found in four study networks. Among proteins, TNF and FGF23 were exploited more and found their involvement in BP and glucose regulation. This is supported by an earlier study, in which the role of TNF-a has been manifested in HTN development through the renin angiotensin aldosterone system (RAAS) pathway in the wild-type mice relative to the TNF-a null mouse models.[57] Besides this, TNF-a triggers oxidative stress, inflammation, HTN, insulin resistance, and the development of type 2 DM by activating nicotinamide adenine dinucleotide phosphate (NADPH) oxidase.[58] On the other hand, the useful role of Vitamin D3 supplementation was reported to downregulate pro-inflammatory cytokines, including TNF-a,[59,60] thus indicating the putative role of Vitamin D3 in BP and glucose regulation.
Recent studies have reported a relationship between increased levels of FGF23 and with incidence of HTN in adult humans. Being an inhibitor, FGF23 overexpression causes a pronounced downfall of 1,25(OH)2D3 level. Vitamin D plays a role in keeping the KL protein level within a permissible range that is required for normal vascular functioning.[61] Decreased levels of KL cause vascular disintegration and endothelial malfunctioning.[35] Other than TNF-a and FGF23, over-expression of GLUT4 channel, a product of SLC2A4 is involved in the downregulation of HTN and prevention of vascular arterial aging.[62]
Networks created in the present study indicated several pathways connected with Vitamin D3 targeted genes. These pathways also seemed to be linked with various genes involving in HTN and diabetes predisposition. In VDR null mice, high systolic BP was studied due to the modulation of the RAAS pathway and vascular endothelial cells’ physiology,[63,64] whereas VDR overexpression in pancreatic b-cells of wild-type mice mitigates DM than the VDR knockout mice.[65] Nuclear receptors meta-pathway regulates solute carrier family proteins participating in BP regulation by enhancing body fluid retention. Furthermore, it modulates PPARa, contributing to cellular metabolism, glucose homeostasis, Ang-II reduction, and Na reabsorption.[66] Vitamin D3 activates signaling molecules such as phospholipase C and phospholipase A2, phosphatidylinositol-3 kinase accompanied by activation of protein kinases, that is, protein kinase A, mitogen-activated protein kinase, protein kinase C, and Ca2+-calmodulin kinase II.[67] Similarly, oxidation by the cytochrome p450 pathway regulates the expression of various cytochrome p450 enzymes such as CYP3A4, CYP2C9, CYP1A1, CYP1A2, CYP17A1, CYP11A1, CYP11B1, and CYP11B2 involved in the modulation of complex biological pathways linked to HTN.
The four Vitamin D3 metabolites and disease-related networks are a knowledge resource and shared through de NDEx https://www.ndexbio.org. These resources can be used by researchers to find a list of genes known to be targeted by Vitamin D3 metabolites and associated with either HTN or DM (Links of networks are given along with figures’ captions). Bioinformaticians can include the genes in their over-representation analysis. Computational biologists can include the Vitamin D3-disease networks in their advanced network analysis approaches.
From the present study, it is recommended to the health practitioner to implement the screening and monitoring of Vitamin D levels of cardiometabolic patients. By knowing the health condition and genetics of an individual, practitioners can incorporate Vitamin D testing, supplementation, and lifestyle interventions into patient care based on patient needs. For patients, taking care of weight, lifestyle management and adopting these strategies supports better management and prevention of conditions such as diabetes, HTN, and CVD.
CONCLUSION
From the present study, it could be concluded that all study subjects including healthy controls and cardiometabolic patients were Vitamin D deficient. However, patients were found to be more Vitamin D deficient than the healthy subjects. VDR polymorphism (rs2228570) and CYP2R1 polymorphism (rs10766197) were significantly linked with HTN and DM, respectively. No association was observed between the studied polymorphisms and serum Vitamin D3 metabolites levels. Furthermore, use of bioinformatics and computational approaches helped to highlight the proteins that seemed to be linked with Vitamin D3 and its related genes, which further interacted and modulated the genes involving the clinical conditions such as cardiovascular disorders, HTN, and DM. Findings of this study can provide guidelines for the further studies to evaluate the role of Vitamin D3 and its related genes/polymorphisms in the pathophysiology of metabolic diseases including CVDs, HTN, and DM. There are some limitations of present study. Vitamin D levels were not assessed for all subjects due to cost. No gene expression data was obtained; hence, the relationship between SNPs and Vitamin D3 related genes could not be investigated. Therefore, more comprehensive studies are required to evaluate the influence of study SNPs on their respective gene expression in Pakistani population. Similarly, effect of VDR, CYP2R1, and CYP27B1 on other genes associated with the complex phenotypes needs to be investigated through further research. Lack of association between the investigated polymorphisms and serum Vitamin D3 level might be due to the ethnicity differences and limited sample size. Future research is warranted to explore the specific impact of polymorphisms (SNPs) on the VDR protein and Vitamin D-synthesizing enzymes’ expression. Moreover, the relationship between polymorphisms in Vitamin D-related genes and serum Vitamin D3 levels needs validation through large-scale cohort studies in the Pakistani population.
Acknowledgement
Authors would like to thank all the participants of this study as well as hospital staff and doctors (especially Dr Shahid Abbas, Dr M Naeem Aslam and Dr Ahmed Bilal) for their help with sample collection. Furthermore, we thank Dr. Haq Nawaz Khan for helpful discussion regarding genotyping assays.
Authors’ contributions
HF and FRA: Conceived this study; HF: Prepared initial draft of manuscript; FRA and ARK: Supervised and proofread the manuscript; SC: Involved in the study design and manuscript drafting; MH: Engaged in all healthy control and patient sampling; HF: Performed the data collection and analysis; FRA, ARK, FE, CTE and SC: Revised the manuscript critically for important intellectual content and all the co-authors finally approved the manuscript.
Ethical approval
Ethical review committee of National Institute for Biotechnology and Genetic Engineering, NIBGE, Faisalabad, Pakistan approved the study. This study was performed in line with the principles of the Declaration of Helsinki.
Declaration of patient consent
Written consent and questionnaire data (i.e., demographic, anthropometric, life-style, dietary habits, health, and family history) were collected from participants.
Financial support and sponsorship
The part of this study is funded by Higher Education Commission (HEC), Pakistan.
Conflicts of interest
There are no conflicts of interest.
Availability of data and materials
The data sets used during the present study are available from the corresponding author on reasonable request.
References
- Vitamin D deficiency and cardiometabolic syndrome: Is the evidence solid? Arch Med Health Sci. 2017;5:229-36.
- [CrossRef] [Google Scholar]
- Global status report on noncommunicable diseases 2014. 2014. Geneva: World Health Organization; Available from: https://www.who.int/publications/i/item/9789241564854
- [Google Scholar]
- Noncommunicable diseases country profiles 2018. 2018. Geneva: World Health Organization; Available from: https://www.who.int/publications/i/item/9789241514620
- [Google Scholar]
- Hypervitaminosis-an emerging pathological condition. Int J Health Sci Res. 2018;8:280.
- [Google Scholar]
- Role of Vitamin D in cardiometabolic diseases. J Diabetes Res. 2013;2013:243934.
- [CrossRef] [PubMed] [Google Scholar]
- Association of serum vitamin D and the risk of cardiovascular diseases among diabetic patients: A systematic review and meta-analysis. Clin Nutr ESPEN. 2024;62:66-75.
- [CrossRef] [PubMed] [Google Scholar]
- Association between vitamin D Status and cardiometabolic risk factors in adults with type 2 diabetes in Shenzhen, China. Front Endocrinol (Lausanne). 2024;15:1346605.
- [CrossRef] [PubMed] [Google Scholar]
- Vitamin D pathway-related gene polymorphisms and their association with metabolic diseases: A literature review. J Diabetes Metab Disord. 2020;19:1701-29.
- [CrossRef] [PubMed] [Google Scholar]
- High-dose vitamin D does not prevent postoperative recurrence of Crohn's disease in a randomized placebo-controlled Trial. Clin Gastroenterol Hepatol. 2021;19:1573-82.e5.
- [CrossRef] [PubMed] [Google Scholar]
- The photobiology of vitamin D and its consequences for humans. Ann N Y Acad Sci. 1985;453:1-13.
- [CrossRef] [PubMed] [Google Scholar]
- Vitamin D metabolism, mechanism of action, and clinical applications. Chem Biol. 2014;21:319-29.
- [CrossRef] [PubMed] [Google Scholar]
- Vitamin D at the intersection of health and disease: The immunomodulatory perspective. Int J Health Sci. 2024;18:1-4.
- [Google Scholar]
- Vitamin D levels and vitamin D receptor genetic variants in egyptian cardiovascular disease patients with and without diabetes. Egypt J Med Hum Genet. 2021;22:55.
- [CrossRef] [Google Scholar]
- Effect of cytochrome P450 family 2 subfamily r member 1 variants on the predisposition of coronary heart disease in the Chinese han population. Front Cardiovasc Med. 2021;8:652729.
- [CrossRef] [PubMed] [Google Scholar]
- A bioinformatics workflow to decipher transcriptomic data from vitamin D Studies. J Steroid Biochem Mol Biol. 2019;189:28-35.
- [CrossRef] [PubMed] [Google Scholar]
- From SNPs to pathways: Biological interpretation of type 2 diabetes (T2DM) genome wide association study (GWAS) results. PLoS One. 2018;13:e0193515.
- [CrossRef] [PubMed] [Google Scholar]
- Serum uric acid: An independent risk factor for cardiovascular disease in pakistani Punjabi patients. BMC Cardiovasc Disord. 2024;24:546.
- [CrossRef] [PubMed] [Google Scholar]
- 2017 ACC/AHA/AAPA/ABC/ACPM/AGS/APhA/ASH/ASPC/NMA/PCNA Guideline for the prevention, detection, evaluation, and management of high blood pressure in adults: A report of the American College of cardiology/American heart association task force on clinical practice guidelines. J Am Coll Cardiol. 2018;71:e127-248.
- [CrossRef] [Google Scholar]
- Intensive blood pressure-lowering strategy should also be offered to older patients. J Hypertens. 2021;39:812-3.
- [CrossRef] [PubMed] [Google Scholar]
- Host and environmental factors influencing individual human cytokine responses. Cell. 2016;167:1111-24.e13.
- [CrossRef] [PubMed] [Google Scholar]
- Determination of human reference values for serum total 1, 25-dihydroxyvitamin D using an extensively validated 2D ID-UPLC-MS/MS method. J Steroid Biochem Mol Biol. 2016;164:127-33.
- [CrossRef] [PubMed] [Google Scholar]
- Association of vitamin D receptor polymorphisms with cardiometabolic conditions in pakistani population. Int J Vitam Nutr Res. 2022;94:45-53.
- [CrossRef] [PubMed] [Google Scholar]
- The ensembl variant effect predictor. Genome Biol. 2016;17:1-14.
- [CrossRef] [PubMed] [Google Scholar]
- Variant effect predictor analysis. Germany: European Bioinformatics Institute (EMBL-EBI) and the Wellcome Sanger Institute; Available from: https://www.ensembl.org/tools/vep [Last accessed on 2025 Mar 21]
- [Google Scholar]
- Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498-504.
- [CrossRef] [PubMed] [Google Scholar]
- Comparative Toxicogenomics database (CTD): Update 2021. Nucleic Acids Res. 2021;49:D1138-D43.
- [CrossRef] [PubMed] [Google Scholar]
- National Institute of Environmental Health Sciences. 2004. Available from: https://ctdbase.org [Last accessed on 2025 Mar 21]
- [Google Scholar]
- DisGeNET: A comprehensive platform integrating information on human disease-associated genes and variants. Nucleic Acids Res. 2017;45:D833-9.
- [CrossRef] [PubMed] [Google Scholar]
- CyTargetLinker app update: A flexible solution for network extension in cytoscape. F1000Res. 2018;7:ELIXIR-743.
- [CrossRef] [Google Scholar]
- Wikipathways Linksets. 2008. Available from: https://cytargetlinker.github.io/pages/linksets [Last accessed on 2025 Mar 21]
- [Google Scholar]
- DisGeNET: A comprehensive platform integrating information on human disease-associated genes and variants. Nucleic Acids Res. 2016;48:D845-55.
- [CrossRef] [PubMed] [Google Scholar]
- Fibroblast growth factor-23, heart failure risk, and renin-angiotensin-aldosterone-system blockade in hypertension: The MESA study. Am J Hypertens. 2019;32:18-25.
- [CrossRef] [PubMed] [Google Scholar]
- Role of klotho in the development of essential hypertension. Hypertension. 2021;77:740-50.
- [CrossRef] [PubMed] [Google Scholar]
- Role of klotho in hyperglycemia: Its levels and effects on fibroblast growth factor receptors, glycolysis, and glomerular filtration. Int J Mol Sci. 2021;22:7867.
- [CrossRef] [PubMed] [Google Scholar]
- The molecular mechanisms by which vitamin D improve glucose homeostasis: A mechanistic review. Life Sci. 2020;244:117305.
- [CrossRef] [PubMed] [Google Scholar]
- Vitamin D Supplementation inhibits oxidative stress and upregulate SIRT1/AMPK/GLUT4 cascade in high glucose-treated 3T3L1 adipocytes and in adipose tissue of high fat diet-fed diabetic mice. Arch Biochem Biophys. 2017;615:22-34.
- [CrossRef] [PubMed] [Google Scholar]
- Vitamin D effects on cell differentiation and stemness in cancer. Cancers (Basel). 2020;12:2413.
- [CrossRef] [PubMed] [Google Scholar]
- Influence of vitamin D status and vitamin D3 Supplementation on genome wide expression of white blood cells: A randomized double-blind clinical trial. PLoS One. 2013;8:e58725.
- [CrossRef] [PubMed] [Google Scholar]
- Single nucleotide polymorphisms in the vitamin D metabolic pathway and their relationship with high blood pressure risk. Int J Mol Sci. 2023;24:5974.
- [CrossRef] [PubMed] [Google Scholar]
- Association of CYP2R1 rs10766197 with MS risk and disease progression. J Neurosci Res. 2018;96:297-304.
- [CrossRef] [PubMed] [Google Scholar]
- Vitamin D level and gene polymorphisms in korean children with type 1 diabetes. Pediatr Diabetes. 2019;20:750-8.
- [CrossRef] [PubMed] [Google Scholar]
- Triangular relationship between single nucleotide polymorphisms in the CYP2R1 gene (rs10741657 and rs12794714), 25-Hydroxyvitamin D levels, and coronary artery disease incidence. Biomarkers. 2014;19:488-92.
- [CrossRef] [PubMed] [Google Scholar]
- Triangular relationship between CYP2R1 gene polymorphism, serum 25 (OH) D3 levels and T2DM in a chinese rural population. Gene. 2018;678:172-6.
- [CrossRef] [PubMed] [Google Scholar]
- Synergism of CYP2R1 and CYP27B1 polymorphisms and susceptibility to type 1 diabetes in egyptian children. Cell Immunol. 2012;279:42-5.
- [CrossRef] [PubMed] [Google Scholar]
- The association of VDR polymorphisms and type 2 diabetes in older people living in community in santiago de chile. Nutr Diabetes. 2018;8:1-9.
- [CrossRef] [PubMed] [Google Scholar]
- Risk conferred by foki polymorphism of vitamin d receptor (VDR) gene for essential hypertension. Indian J Hum Genet. 2011;17:201-6.
- [CrossRef] [PubMed] [Google Scholar]
- Fok I and Bsm I gene polymorphism of vitamin D receptor and essential hypertension: A mechanistic link. J Clin Hypertens. 2023;29:5.
- [CrossRef] [PubMed] [Google Scholar]
- Study of vitamin D receptor gene polymorphisms in a cohort of myocardial infarction patients with coronary artery disease. BMC Cardiovasc Disord. 2021;21:1-9.
- [CrossRef] [PubMed] [Google Scholar]
- Vitamin D receptor (VDR) gene polymorphisms and risk of coronary artery disease (CAD): Systematic review and meta-analysis. Biochem Genet. 2021;59:813-36.
- [CrossRef] [PubMed] [Google Scholar]
- Vitamin D and cardiovascular diseases: From physiology to pathophysiology and outcomes. Biomedicines. 2024;12:768.
- [CrossRef] [PubMed] [Google Scholar]
- VDBP, CYP27B1, and 25-hydroxyvitamin D gene polymorphism analyses in a group of sicilian multiple sclerosis patients. Biochem Genet. 2017;55:183-92.
- [CrossRef] [PubMed] [Google Scholar]
- Association between polymorphisms in vitamin D pathway-related genes, vitamin D status, muscle mass and function: A systematic review. Nutrients. 2021;13:3109.
- [CrossRef] [PubMed] [Google Scholar]
- Strong association between VDR FokI (rs2228570) gene variant and serum vitamin D levels in turkish cypriots. Mol Biol Rep. 2019;46:3349-55.
- [CrossRef] [PubMed] [Google Scholar]
- The study of the association between rs2228570 polymorphism of VDR gene and vitamin D blood serum concentration in the inhabitants of the russian arctic. Voprosy Istorii. 2017;86:77-84.
- [Google Scholar]
- Involvement of tumor necrosis factor-alpha in angiotensin ii-mediated effects on salt appetite, hypertension, and cardiac hypertrophy. Hypertension. 2008;51:1345-51.
- [CrossRef] [PubMed] [Google Scholar]
- TNF-α in the cardiovascular system: From physiology to therapy. Int J Interferon Cytokine Mediat Res. 2015;7:9-25.
- [CrossRef] [Google Scholar]
- Association between levels of vitamin D and inflammatory markers in healthy women. J Inflamm Res. 2016;9:51.
- [CrossRef] [PubMed] [Google Scholar]
- Vitamin D and immune regulation: Antibacterial, antiviral, anti-inflammatory. JBMR Plus. 2020;5:e10405.
- [CrossRef] [PubMed] [Google Scholar]
- Effect of vitamin D Supplementation on klotho protein, antioxidant status and nitric oxide in the elderly: A randomized, double-blinded, placebo-controlled clinical trial. Eur J Integr Med. 2020;35:101089.
- [CrossRef] [Google Scholar]
- Maintenance of GLUT4 expression in smooth muscle prevents hypertension-induced changes in vascular reactivity. Physiol Rep. 2015;3:e12299.
- [CrossRef] [PubMed] [Google Scholar]
- Vitamin D receptor deficiency increases systolic blood pressure by upregulating the reninangiotensin system and autophagy. Exp Ther Med. 2022;23:1-8.
- [CrossRef] [PubMed] [Google Scholar]
- Elimination of vitamin D receptor in vascular endothelial cells alters vascular function. Hypertension. 2014;64:1290-8.
- [CrossRef] [PubMed] [Google Scholar]
- Vitamin D receptor overexpression in β-cells ameliorates diabetes in mice. Diabetes. 2020;69:927-39.
- [CrossRef] [PubMed] [Google Scholar]
- Impact of peroxisome proliferator-activated receptor-α on diabetic cardiomyopathy. Cardiovasc Diabetol. 2021;20:2.
- [CrossRef] [PubMed] [Google Scholar]








